INTRODUCTION
Since the emergence of COVID-19 pandemic, which was caused by SARS-CoV-2, the impact on global public health and the economy has been severe [1]. In addition, numerous variants of concern (VOCs) have arisen [2]. The dominant VOCs have been Omicron descendent lineages since 2023. Currently, the variants of interest (VOIs) under WHO tracking are XBB.1.5, XBB.1.16, EG.5, BA.2.86, and JN.1. Omicron sub-lineages exhibit significant immune evasion from all vaccines and most of the therapeutic drugs on the market prior to 2023. The level of neutralization antibodies against Omicron variants is significantly reduced based on analyses of serum samples from people who received different types of vaccines [3–6]. These findings suggest an elevated risk of infection.
Several types of COVID-19 vaccines have been developed, including inactivated, live-attenuated, recombinant subunit, vector-based, and nucleic acid vaccines [7–12]. Among the vaccines, the messenger RNA (mRNA) vaccine is regarded as the most promising pandemic-response vaccine because the mRNA vaccine has a flexible immunogen design, can be manufactured rapidly, and is scalable [13,14]. BNT162b2 and mRNA-1273 were the first two mRNA vaccines that were market-approved against SARS-CoV-2 [14–16]. Both BNT162b2 and mRNA-1273 include mRNA that encodes the SARS-CoV-2 spike (S) protein. The receptor-binding domain on the virus binds to the angiotensin-converting enzyme 2 (ACE2) receptor on the cells of the host [17–22]. Omicron variants have several mutations in the S protein compared to the original virus, which leads to decreased neutralization antibodies against these variants [3]. There are several ways to prevent new VOCs infections. Researchers focus on designing new vaccines based on the variants for broad-spectrum virus prevention [23–25]. However, the entire process from vaccine design-to-market approval is long. During this lengthy process, several new mutations of the virus possibly emerge. We should also notice that many people have been dosed with vaccines designed based on the original virus. Some studies have demonstrated that primary and booster vaccinations with the marketed vaccines prevent severe COVID-19 and death [26–28]. Thus, it is still meaningful to conduct further studies to evaluate the value of vaccines developed from the ancestral strain.
In this study we developed an mRNA vaccine (codename: LVRNA009) expressing the S glycoprotein and evaluated its immunogenicity. Eight months after the primary and one boost immunization, LVRNA009 was shown to elicit neutralization antibodies that provided protection against both the Wuhan-Hu-1 strain and Omicron variants of SARS-CoV-2. Furthermore, the 2nd LVRNA009 boost induced much higher neutralization antibody titers against SARS-CoV-2 variants and protected the mice from Omicron variant challenge. Moreover, clinical studies have shown that LVRNA009 has good safety and efficacy. LVRNA009 provides long-term protection against SARS-CoV-2 variants and confers better protection with another booster dose. These findings indicate that LVRNA009, a vaccine designed based on the original virus strain, might be effective in management of the COVID-19 pandemic.
MATERIALS AND METHODS
Chemicals, cytokines, and peptides
A set of 20-mer peptides encompassing the SARS-CoV-2 S protein and overlapping by 10 amino acids was synthesized (GL Biochem Ltd., Shanghai, China) and used for T cell stimulation.
Vaccine design and production
The SARS-CoV-2 vaccines were designed with mRNA encoding the Wuhan-Hu-1 S protein (Fig 1A). The vaccine was produced based on the Liverna Therapeutics platform (China patent: ZL201911042634.2), as previously described [29].
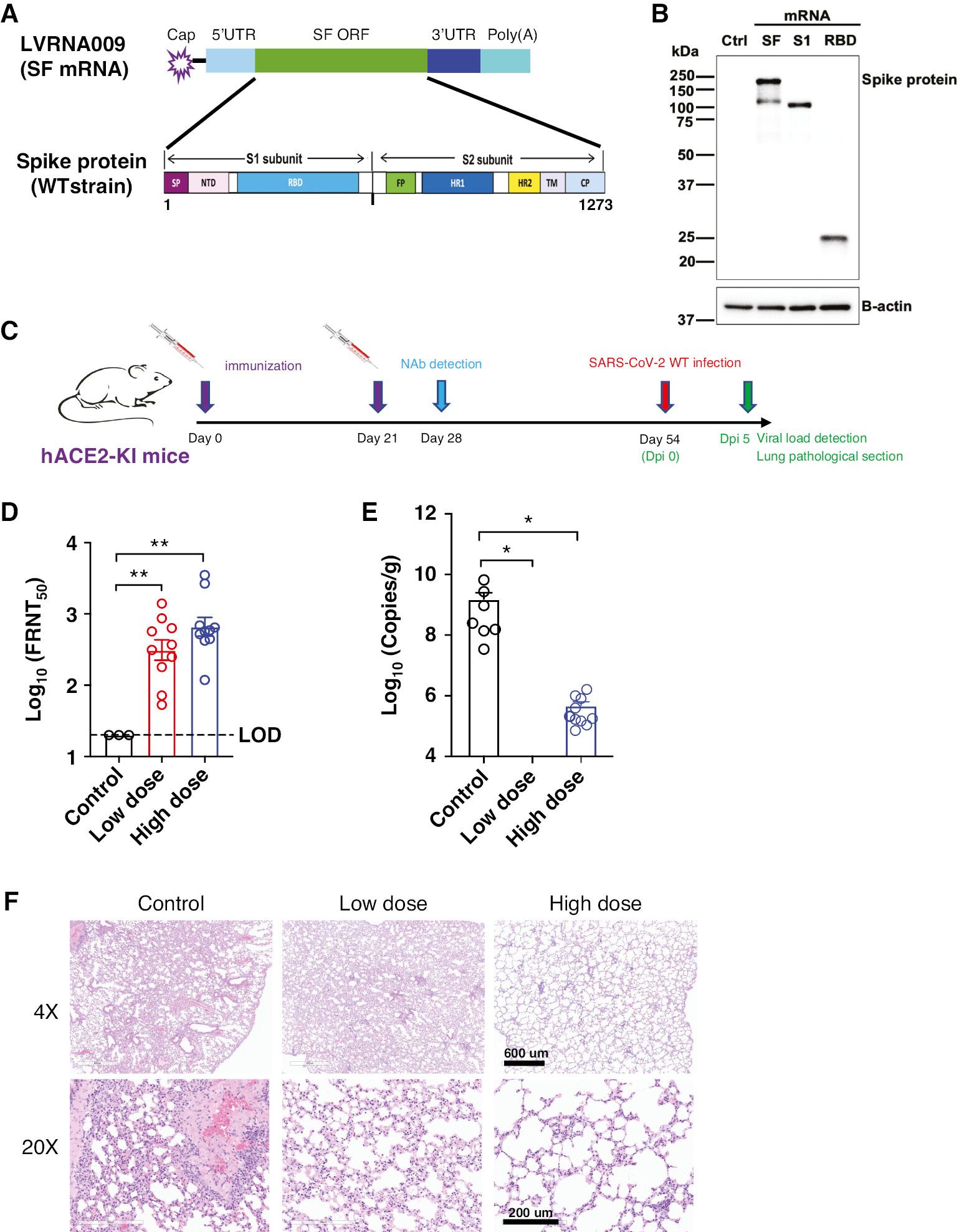
Design, characterization and immunogenicity evaluation of the mRNA vaccine against SARS-CoV-2. (A) Schematic diagram of the target antigen-Spike (S) protein, encoded by mRNA vaccines against the Wuhan-Hu-1. (B) The expression of S, S1 or RBD proteins in HEK293 cells transfected with in vitro transcribed mRNA was detected by western blot. (C) Animal experimental strategy. (D) The neutralizing titers against SARS-CoV-2 in animals were tested by FRNT. hACE2-KI mice, N=10. *P<0.05, **P<0.01. (E) The Viral loads of SARS-CoV-2 in animals were tested by qPCR. (F) H&E staining was used to examine histopathologic changes in the lung tissues. Scale bar, 600 μm (4×) or 200 μm (20×).
mRNA transfection
HEK293 cells were cultured in DMEM supplemented with penicillin/streptomycin and 10% FBS (Gibco, Grand Island, NY, USA). Transfection was performed with Lipofectamine® MessengerMAX™ Transfection Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. After transfection, the cells were incubated under specific conditions for 24 h prior to harvesting.
Detection of vaccine antigens using western blot analysis
HEK293 cells were harvested and lysed post-transfection. The protein extracts underwent separation via 12% SDS-PAGE gel and were then transferred onto PVDF membranes. Following a 1-h blocking step, the membranes were probed with anti-SARS-CoV-2 S glycoprotein monoclonal antibody (1:2000; Sino Biological, Beijing, China) and β-actin (1:10,000; Sigma-Aldrich, St. Louis, MO, USA) at ambient temperature for 60 min. A secondary antibody HRP-conjugated goat anti-mouse IgG antibody (1:5000; BBI Life Sciences, Cambridge, UK) was then applied for 1 h. Visualization of the immunoreactive bands was achieved using enhanced chemiluminescence substrate (Perkin-Elmer, Waltham, MA, USA) with imaging using the ChemiDoc XRS+ system (Bio-Rad, Hercules, CA, USA).
Viruses
The SARS-CoV-2 variants, including WT (GenBank: MT123291), Omicron BA.1, and BA.2.3 (IQTC-IM22003633), were isolated from COVID-19 patients on Vero E6 cells. Virus information of the original sampling is displayed in S1 Table. BA.5 (GDPCC-303) virus was donated by the Guangdong Provincial Center for Disease Control and Prevention (Guangdong, China). The complete genomic sequence was acquired using next-generation sequencing techniques. SARS-CoV-2 variants genomic sequences were assigned to “Nextstrain clades” using the Nextclade tool (https://clades.nextstrain.org). These viruses are preserved in the Guangzhou Customs District Technology Center BSL-3 Laboratory (Guangdong, China).
Animal experiments and approvals
BALB/c mice (6-8 weeks old) were purchased from the Hunan SJA Laboratory Animal Co. (Hunan, China). The hACE2-KI/NIFDC mouse model was established and supplied by the Institute for Laboratory Animal Resources, NIFDC (Beijing, China) [30].
The hACE2-KI mice were divided into groups (N=10/group) and given doses (5 [low dose] or 15 μg [high dose]) for immunization on days 0 and 21. Serum samples were collected 28 days after the first immunization to determine the anti-SARS-CoV-2 virus neutralization titers. The mice were infected with SARS-CoV-2 (5×104 focus-formation units [FFU]) using the Wuhan-Hu-1 strain following immunization. All mice were euthanized 5 days post-infection and lung tissues were extracted for examination of histopathologic changes and determination of virus titers.
BALB/c mice (N=3–4/group) were used to evaluate the T cell response induced by the mRNA vaccine. The animals were divided into groups and given doses (5 [low dose] or 15 μg [high dose]) for immunization on days 0 and 21. Spleens of 3 mice per group were collected on days 7, 21, 28, and 42. The remaining mice were transduced with Ad5-hACE2 (2.5×108 FFU/75 μL) intranasally on day 58. These mice were infected with SARS-CoV-2 (5×104 FFU) using the Wuhan-Hu-1 strain following transduction. On days 3, 5, and 8 post-infection, 3–4 mice per group were euthanized and the spleens and bronchoalveolar lung fluid (BALF) were harvested and prepared for flow cytometry analysis.
BALB/c mice (N=6–7/group) were used to evaluate protection from SARS-CoV-2 variants by LVRNA009. The animals were divided into groups and given doses (15 μg) for immunization on days 0 and 21. Serum samples were then collected 42 and 261 days after the first immunization to determine the anti-SARS-CoV-2 virus neutralization titers. On day 291, The mice were infected with SARS-CoV-2 (5×104 FFU) using Wuhan-Hu-1 or Omicron BA.1.1 strain following transduction. All mice were euthanized and lung tissues were harvested for detection of virus titers 2 days post-infection.
The mouse studies were approved by Animal Care and Use Form of Affiliated First Hospital of Guangzhou Medical University (2021059) and performed in accordance with the Guide for the Care and Use of Laboratory Animals. All work with SARS-CoV-2 was performed in the Biosafety Level 3 (BSL-3) Laboratory of Guangzhou Customs District Technology Center.
Focus-forming assay (FFA) for SARS-CoV-2
Vero E6 cells were cultured in 96-well plates with DMEM containing 10% FBS at 37°C in a 5% CO2 atmosphere. The virus or supernatants from homogenized mouse lungs were serially diluted by DMEM. The diluted samples were then introduced to Vero E6 cells in 96-well plates and incubated at 37°C in a 5% CO2 environment for 60 min. A 1.6% carboxymethylcellulose (CMC) layer was applied to the plates following removal of the inoculum. After a 24-h incubation period with the virus, cells were fixed and permeabilized with 4% paraformaldehyde and 0.2% Triton X-100, respectively. The cells were subsequently treated with a rabbit polyclonal antibody against the SARS-CoV-2 nucleocapsid protein (Sino Biological), followed by a goat anti-rabbit secondary antibody labelled with HRP (Jackson ImmunoResearch, West Grove, PA, USA). The foci were visualized using TrueBlue reagent and quantified with an ELISPOT reader (Cellular Technology Limited, Cleveland, OH, USA). The viral titers were determined as FFU per milliliter or gram.
Focus reduction neutralization test (FRNT) for SARS-CoV-2
FRNTs were performed as previously described [31]. Serum samples were serially diluted with DMEM and mixed with SARS-CoV-2 containing 150–200 FFU at a 1:1 ratio (v/v). After a 1-h incubation at 37°C, aliquots were introduced to Vero E6 cells in 96-well plates and incubated for an additional 1 h. Following removal of the inoculum, a 1.6% CMC layer was applied to the plates. The cells were then fixed with 4% paraformaldehyde 24 h after infection. The plates were stained and counted using the FFA method. FRNT50 were calculated using the 4PL regression model.
Detection of SARS-CoV-2 viral load by qPCR
Lung tissues were homogenized with TRIzol (Invitrogen), then RNAs were extracted using a Universal RNA Purification kit (EZBioscience, Roseville, MN, USA) according to the manufacturer’s instructions. The virus load was detected by qPCR using the Detection Kit for 2019-nCoV RNA (Daangene, Guangzhou, China). The pUC57-2019-nCoV-N plasmid (Genscript, Nanjing, China) was used as a control and virus loads are expressed as copies per gram of tissue.
Histopathologic examination
Lung tissues were harvested and preserved in a 10% formaldehyde solution. The tissue samples were then dehydrated using a sequential ethanol gradient, embedded in paraffin, and sliced into 4-μm thick sections. These sections were subsequently deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E). Finally, the stained sections were examined and photographed under a light microscope.
Preparation of peripheral blood mononuclear cells (PBMCs)
Isolation of human PBMCs was performed as detailed in a previous study [32].
Flow cytometry analysis
The following monoclonal anti-mouse antibodies were used: CD4 (cloneRM4-5, Cat. No.: 48-0042-82 eBioscience, San Diego, CA, USA), CD8α (clone 53-6.7, Cat. No.: 100723, Biolegend, San Diego, CA, USA), interferon-γ (IFN-γ) (cloneXMG1.2, Cat. No.: 17-7311-82, eBioscience), tumor necrosis factor (TNF) (cloneMP6-XT22, Cat. No.: 12-7321-82, eBioscience), CD16/32 (2.4G2, BD Biosciences, Franklin Lakes, NJ, USA). Lymphocytes were cultured in 96-well round bottom plate at 37°C for 5-6 h in the presence of 2 μM peptide pool and brefeldin A (BD Biosciences) for intracellular cytokine staining (ICS). The splenocytes suspensions and BALF cells were labeled for cell surface markers, fixed and permeabilized with Cytofix/Cytoperm Solution (BD Biosciences), and labeled with intracellular antibodies. All flow cytometry data were acquired on a BD FACSVerse and analyzed using FlowJo software.
Clinical studies
In this single-center, blinded, placebo-controlled phase II trial, 420 healthy adults (18–59 years of age) received LVRNA009 (50 μg or 100 μg of the study vaccine or placebo). The safety and immunogenicity of LVRNA009 were accessed.
Statistical analysis
Group variances were evaluated by ANOVA utilizing GraphPad Prism 8.0 software (GraphPad Software Inc., San Diego, CA, USA). Statistical significance was denoted by P≤0.05 (*P≤0.05; **P≤0.01; ***P≤0.001; and ****P≤0.0001). All results are presented as the mean ± SEM and adjusted for multiple comparisons.
Institutional review board and informed consent statement
The clinical study was performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Hunan Provincial Center for Disease Control and Prevention (protocol code: LVRNA009-II-01; approved on 28 January 2023). Informed consent was obtained from all subjects involved in the study.
RESULTS
Design and expression of the mRNA vaccines against different SARS-CoV-2 variants
The SARS-CoV-2 S protein is a key target for neutralization antibodies [17]. An mRNA construct (SF mRNA) encoding full-length SARS-CoV-2 Wuhan-Hu-1 strain S protein was developed, as shown in Fig 1A. The nucleotide sequences of this construct were optimized. Using a similar strategy, mRNA encoding the S protein S1 or receptor-binding domain (RBD) subunit was also synthesized. Western blot results revealed that LVRNA009 mRNA (SF mRNA) induced the expression of S and S1 proteins (Fig 1B), which reflect the full-length and cleaved proteins, respectively. The in vitro expression of S1 and RBD proteins induced by S1 and RBD mRNA, respectively, was also confirmed. To obtain vaccine candidates against Wuhan-Hu-1 strain, SF, S1, and RBD mRNA were encapsulated into LNPs, respectively. Higher neutralization antibodies were induced by SF mRNA-LNP than S1 mRNA-LNP and RBD mRNA-LNP in mice (data not shown). Thus, SF mRNA-LNP (also designated LVRNA009) was chosen as the potential mRNA vaccine for the further studies.
To assess the effectiveness of LVRNA009 in protection against SARS-CoV-2, hACE2-KI mice were injected intramuscularly with 2 doses of the vaccine (low [5 μg] or high dose [15 μg] of LVRNA009) on days 0 and 28. The control group received an intramuscular injection of saline (Fig 1C). The VNTs of sera obtained on day 21 from the vaccinated mice were shown to be higher than the control group (Fig 1D). Twenty-six days after the 2nd immunization (day 54), the mice were exposed to a challenge dose of 5×104 FFU of SARS-CoV-2 Wuhan-Hu-1. Results from qPCR analysis revealed a significant decrease in virus copies in lung tissues from mice in the low- and high-dose groups compared to the negative control group (Fig 1E). The control group exhibited severe histopathologic changes in the lung tissues. The Wuhan-Hu-1 vaccine attenuated these histopathologic changes in all vaccine immunization groups (Fig 1F). These findings indicated that LVRNA009 effectively inhibits viral replication and provides hACE2-KI mice strong protection against SARS-CoV-2 infection.
LVRNA009 vaccination induced a robust T cell response in mice
The cellular immune responses in Ad5-hACE2 mice vaccinated with LVRNA009 (high or low dose) were detected and characterized (Fig 2). BALB/c mice were immunized on days 0 and 21 by intramuscular injection. Thirty-nine days after the boosting vaccination the mice were transduced intranasally with adenovirus expressing human ACE2 (Ad5-hACE2) [31], then infected with wild type (WT) SARS-CoV-2 virus after 5 days. Lymphocytes were collected from the spleens of mice to detect T cell responses at multiple time points, as indicated in Fig 2A. When stimulated with the S1 peptide pool, a higher frequency of antigen-specific CD4+ T cells was detected in the high-dose immunization group compared to the control group 21 days after the boost (P<0.01; Fig 2B [top left panel]). Additionally, a significant increase in the frequency (P<0.01) and number (P<0.05) of antigen-specific CD4+ T cells was detected in the high-dose immunization group compared to the control group 7 days post-boost and 8 days post-infection [dpi] (Fig 2B [right panels]).
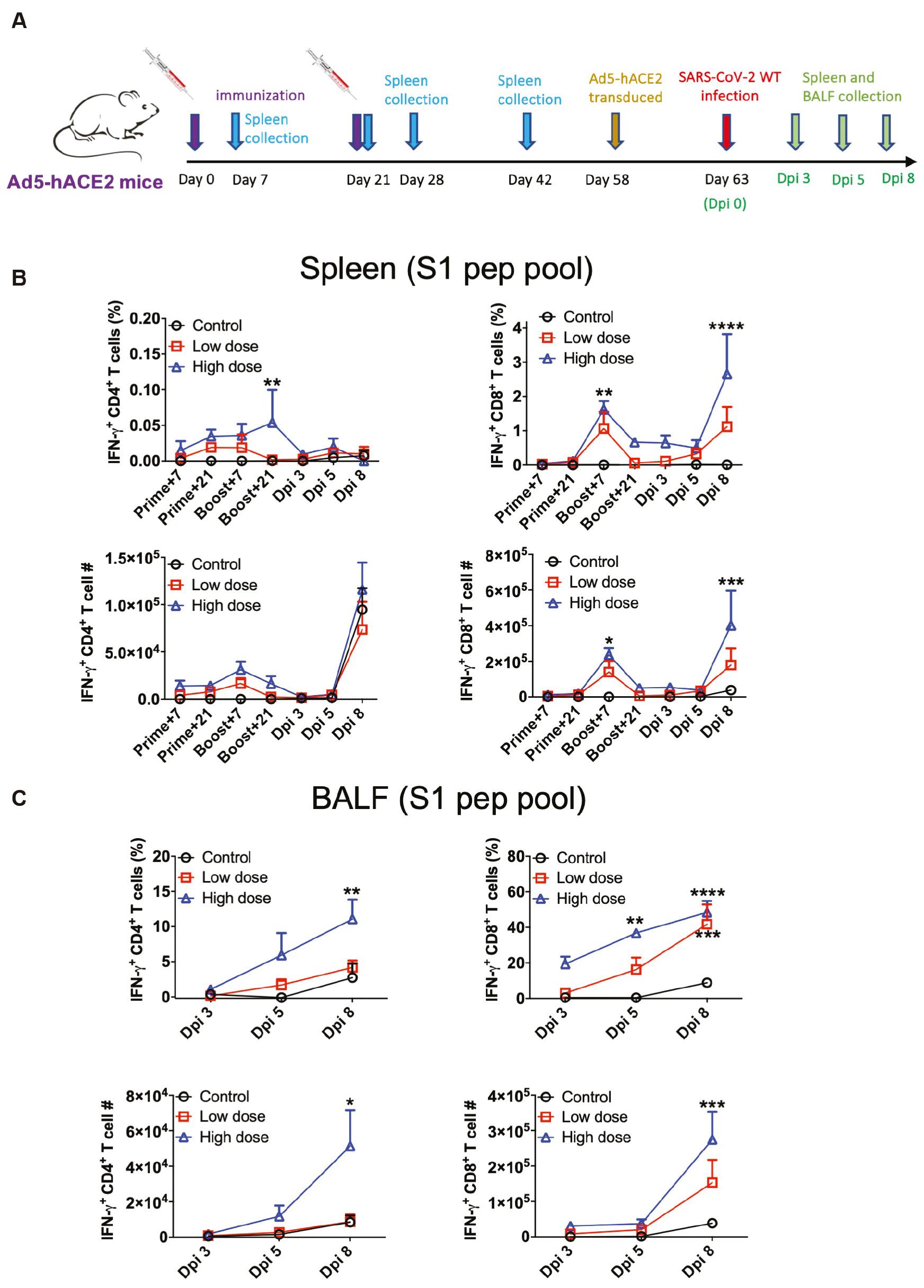
The robust T cell response was induced by LVRNA009 vaccination. (A) Six-week-old female BALB/c mice were immunized with LVRNA009 (5 [low dose] or 15 μg [high dose]) at days 0 and 21 through intramuscular injection. Thirty-nine days after the boosting vaccination, the mice were transduced with 2.5×108 FFU adenovirus expressing human ACE2 (Ad5-hACE2) intranasally, then infected with 5×104 FFU/50 μl WT SARS-CoV-2 virus after 5 days. T cell responses were detected at indicated time points. (B) Lymphocytes from spleens of vaccinated or transduced/infected mice were harvested at indicated time points and stimulated with the S1 peptide pool and for 6 h in the presence of brefeldin A. The frequencies (Top left) and cell numbers (Bottom left) of antigen-specific CD4+ T cells in the spleen are shown (N= 3–4). The frequencies (Top right) and cell numbers (Bottom right) of antigen-specific CD8+ T cells in the spleen are shown (N=3–4). (C) Lymphocytes from BALF of vaccinated or transduced/infected WT BALB/c mice were harvested at indicated time points and stimulated with the S1 peptide pool. The percentage of IFN-γ+ CD4+ T cells (top left), the percentage of IFN-γ+ CD8+ T cells (top right), the number of IFN-γ+ CD4+ T cells (bottom left) and the number of IFN-γ+ CD8+ T cells (bottom right) were shown respectively. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
To examine cellular responses in lungs upon virus challenge, lymphocytes from the BALF of transduced and infected mice were harvested and stimulated with the S1 peptide pool. A significant increased frequency and number of antigen-specific CD4+ T cells was observed in the high-dose immunization group compared to the control group at 8 dpi after peptide stimulation (Fig 2C [left panels]). Similar increasing trends were also observed in antigen-specific CD8+ T cells at 5 and 8 dpi (Fig 2C [right panels]). Taken together, these results suggest that LVRNA009 immunization induces robust Th1 cell responses, which may contribute to memory effects and confer long-term protection against SARS-CoV-2.
LVRNA009 immunization exerts long-term protection in mice and the VNTs against Omicron variants are enhanced by a 2nd boost immunization
BALB/c mice that received LVRNA009 prime and boost immunizations of were maintained for 8 months to evaluate the long-term protection ability of LVRNA009. Then, the mice split into 3 groups (Fig 3A). A group of mice were immunized with 1 additional dose of LVRNA009 and 1 month later the mice were infected with Omicron BA.1.1 virus. Two groups of mice were infected with WT or the Omicron BA.1.1 variant strain without a 2nd vaccine boost. The two groups of mice immunized with saline and infected with WT or Omicron BA.1.1 variant strain were designated as negative controls. Two days post-infection the lungs were collected to determine the viral load (Fig 3B). No live virus was detected in the mice 8 months after the 1st LVRNA009 boost, whether infected with WT or Omicron strain virus. As expected, the live virus was also not detected in the mice with the LVRNA009 2nd boost. In contrast, the mice in the control groups had high live virus titers in the lungs. These results showed that LVRNA009 immunization inhibits virus replication in mice and therefore confer long-term protection against WT and Omicron virus in mice.
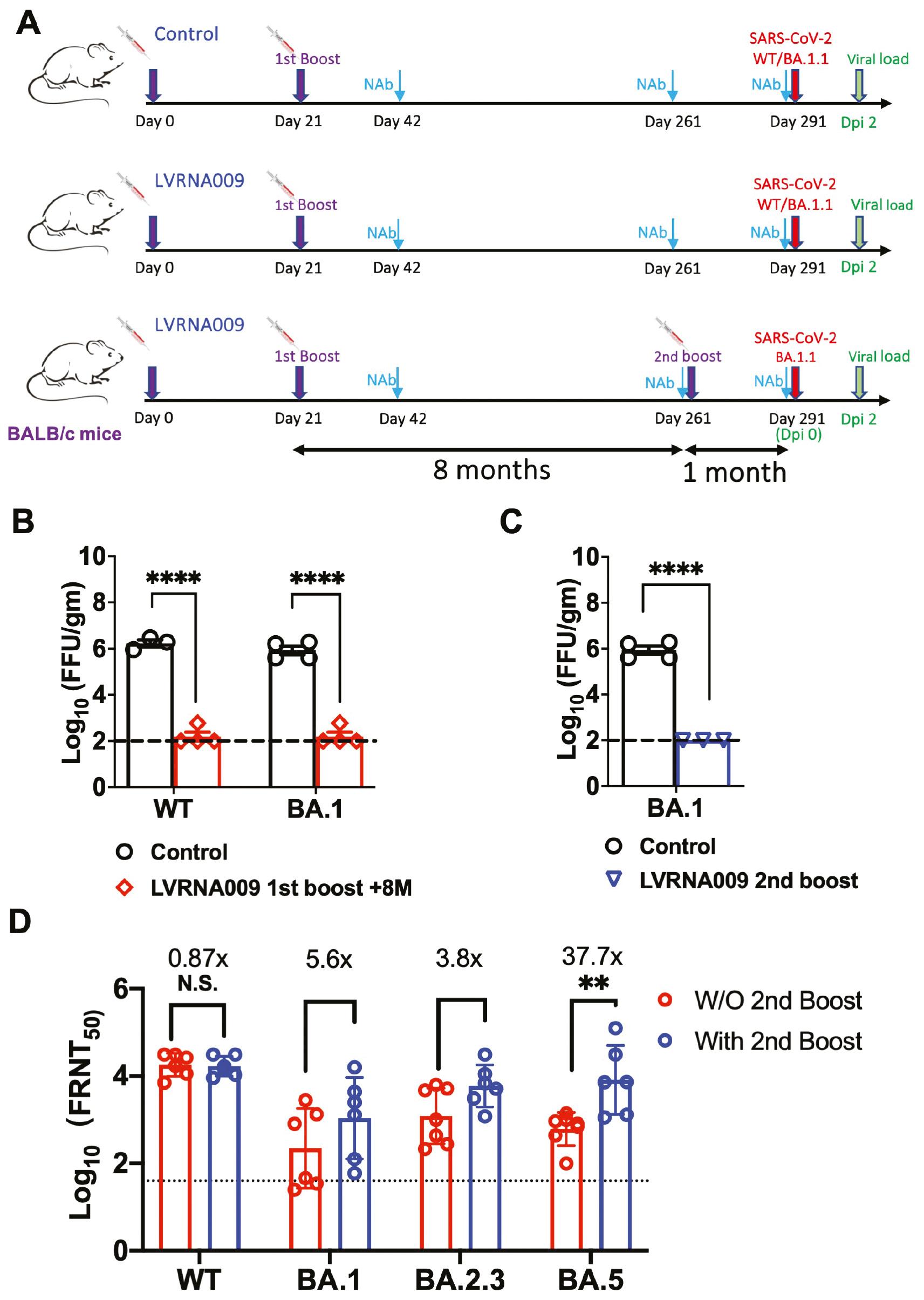
Protection from SARS-CoV-2 variants in mice before and after the 2nd LVRNA009 boost. (A) The schematic diagram of the immunization and virus challenging procedures in WT BALB/c mice. (B) Control mice and LVRNA009 vaccinated mice 8 months after the 1st boost vaccination were challenged with SARS-CoV-2 variants. Viral loads in lungs were detected at 2 dpi. (C) Control mice and LVRNA009 vaccinated mice after the 2nd boost vaccination were challenged with SARS-CoV-2 BA.1.1. Viral loads in lungs were detected at 2 dpi. (D) The VNTs against live SARS-CoV-2 virus variants in mice at indicated time points (N=6–7). N.S. no significant, **P<0.01, ****P<0.0001.
To evaluate the cross-protection effects of LVRNA009, the sera of mice before and after the 2nd boost were collected and the VNTs against WT and several VOCs were detected. High VNTs against the WT strain were detected in mice 8 months after the 1st boost and maintained after the 2nd boost. Compared to VNTs against the WT strain, the VNTs against Omicron variants were decreased to some extent in mice 8 months after the 1st boost. However, the VNTs against all of the VOCs were increased upon the 2nd boost, especially for the VNTs against Omicron BA.5 strain (Fig 3C). These results suggest that one additional booster is important for enhanced protection against the current SARS-CoV-2 VOCs.
LVRNA009 has a safe profile based on a phase II clinical study
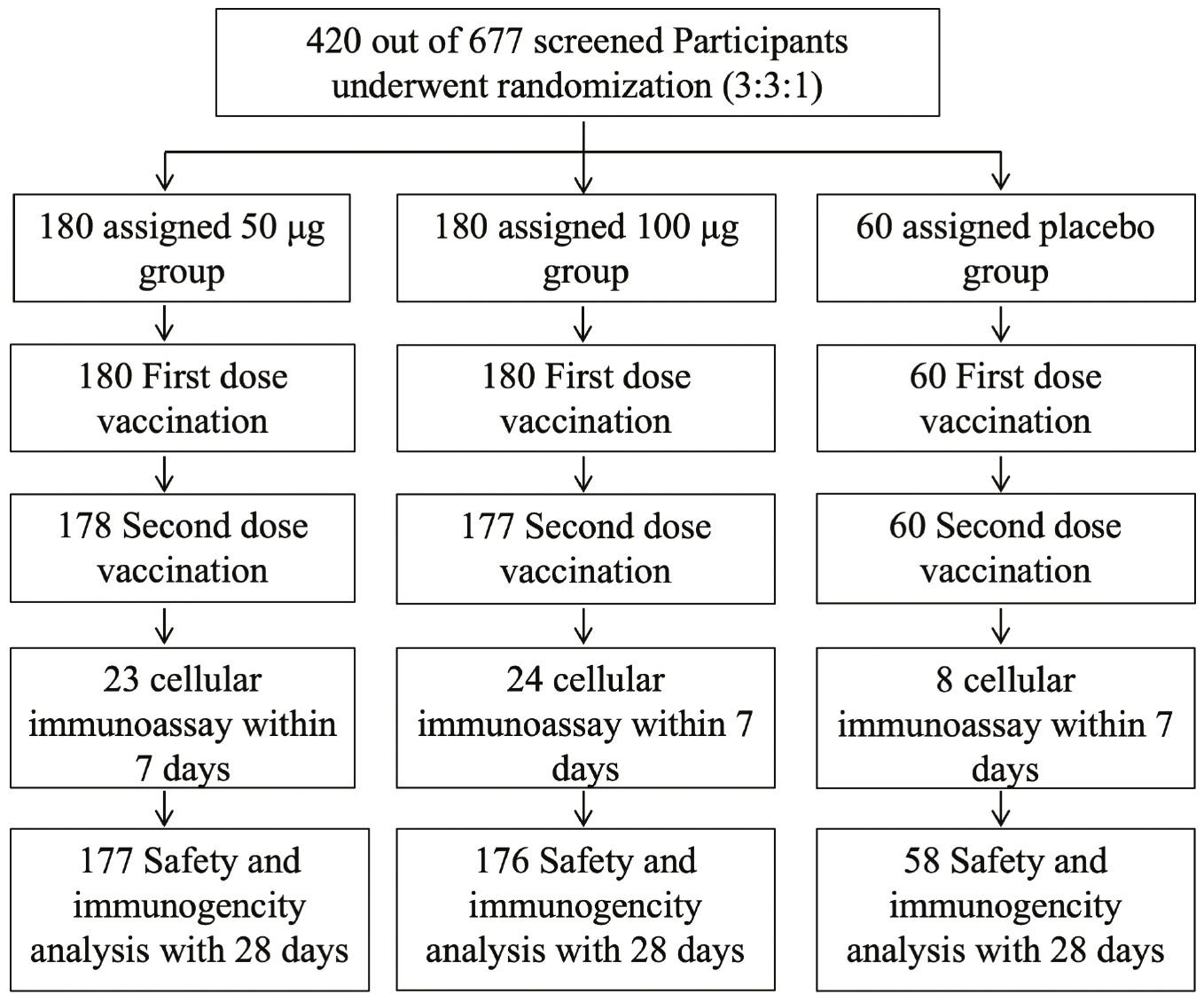
A phase I clinical trial in China involving the LVRNA009 mRNA vaccine has been conducted (Clinicaltrials.gov NCT05364047 and Chictr.org.cn ChiCTR2100049349). Seventy-two eligible participants were recruited and randomized for this clinical study. LVRNA009 demonstrated encouraging safety and tolerability results at all three dose levels in adults [33]. To further evaluate the safety and immunogenicity of LVRNA009 in healthy Chinese adults, a phase II clinical trial was conducted (Clinicaltrials.gov NCT05364047 and Chictr.org.cn ChiCTR2200057782). In this single-center, blinded, placebo-controlled phase II trial, 420 healthy adults (18-59 years of age) received LVRNA009 (50 or 100 μg of the study vaccine or placebo), and the safety and immunogenicity of LVRNA009 were accessed. The entire trial profile is shown in Fig 4.
Eligible volunteers were randomly distributed among the three dose groups, with no group differences in vital signs and demographic characteristics detected prior to vaccination. The demographic characteristics and the distribution are detailed in Table 1.
Demographic characteristics of volunteers.
| 50 μg group (N=180) | 100 μg group (N=180) | Placebo (N=60) | Total (N=420) | Methodology | Value | P value | |
|---|---|---|---|---|---|---|---|
| Age (years) a | |||||||
| n (Missing) | 180 (0) | 180 (0) | 60 (0) | 420 (0) | |||
| Mean (SD) | 38.40 (11.08) | 38.34 (10.74) | 38.85 (10.29) | 38.44 (10.80) | ANOVA | 0.052 | 0.9496 |
| Median | 38.25 | 36.25 | 37.75 | 37.75 | |||
| Min, Max | 19.8, 59.2 | 19.6, 59.7 | 22.0, 58.5 | 19.6, 59.7 | |||
| Gender | |||||||
| Male n (%) | 73 (40.56) | 76 (42.22) | 24 (40.00) | 173 (41.19) | chi-square test | 0.144 | 0.9305 |
| Female n (%) | 107 (59.44) | 104 (57.78) | 36 (60.00) | 247 (58.81) | |||
| Total (missing) | 180 (0) | 180 (0) | 60 (0) | 420 (0) | |||
| Ethnicity | |||||||
| Han n (%) | 175 (97.22) | 179 (99.44) | 59 (98.33) | 413 (98.33) | Fisher’s exact test | NA | 0.2645 |
| Other n (%) | 5 (2.78) | 1 (0.56) | 1 (1.67) | 7 (1.67) | |||
| Total (missing) | 180 (0) | 180 (0) | 60 (0) | 420 (0) | |||
| Weight (kg) | |||||||
| n (missing) | 180 (0) | 180 (0) | 60 (0) | 420 (0) | |||
| Mean (SD) | 61.58 (10.80) | 62.21 (10.80) | 60.47 (10.98) | 61.69 (10.82) | ANOVA | 0.595 | 0.5519 |
| Median | 61.30 | 61.00 | 57.50 | 61.00 | |||
| Min, Max | 39.2, 95.0 | 40.0, 94.0 | 41.0, 94.9 | 39.2, 95.0 | |||
| Height (cm) | |||||||
| n (missing) | 180 (0) | 180 (0) | 60 (0) | 420 (0) | |||
| Mean (SD) | 160.7 (7.4) | 161.7 (8.3) | 159.6 (7.2) | 161.0 (7.8) | ANOVA | 1.900 | 0.1509 |
| Median | 160.0 | 160.5 | 160.0 | 160.0 | |||
| Min, Max | 142, 181 | 145, 183 | 141, 174 | 141, 183 | |||
| BMI (kg/m2) | |||||||
| n (missing) | 180 (0) | 180 (0) | 60 (0) | 420 (0) | |||
| Mean (SD) | 23.791 (3.558) | 23.742 (3.461) | 23.697 (3.642) | 23.757 (3.521) | ANOVA | 0.019 | 0.9812 |
| Median | 23.340 | 23.310 | 23.275 | 23.310 | |||
| Min, Max | 15.51, 33.27 | 16.44, 37.18 | 16.85, 33.15 | 15.51, 37.18 | |||
aAge (years) = (date of informed consent signature - date of birth + 1)/365.25.
The incidence of adverse events (AEs) from the 1st dose to 28 days after full immunization was 78.89%, 81.11%, and 21.67% in the 50 μg, 100 μg, and placebo groups, respectively. The incidence of AEs associated with the study vaccine was 74.44%, 79.44%, and 10.00%, respectively. The incidence of severe AEs (≥ grade 3) was 2.22%, 6.67%, and 0.00%, respectively. The incidence of severe adverse events (SAEs) from the 1st dose to 28 days after full immunization was 1.11%, 0.56%, and 0.00% in the 50 μg, 100 μg, and placebo groups, respectively. These SAEs were unrelated to vaccination according to the study investigator. No AEs of special concern or SAEs leading to death or were life-threatening were observed (Table 2).
Overall occurrence of adverse events (AEs) from the first dose to 28 days after full immunization.
| Adverse events - no. (%) | 50 μg group (N=180) | 100 μg group (N=180) | Placebo group (N=60) | P value |
|---|---|---|---|---|
| All AEs | 142 (78.89) | 146 (81.11) | 13 (21.67) | <0.0001 |
| Related adverse events | 134 (74.44) | 143 (79.44) | 6 (10.00) | <0.0001 |
| AEs of severity ≥ grade 3 | 6 (3.33) | 14 (7.78) | 1 (1.67) | 0.0853 |
| Related AEs of severity ≥ grade 3 | 4 (2.22) | 12 (6.67) | 0 (0.00) | 0.0299 |
| SAEs | 2 (1.11) | 1 (0.56) | 0 (0) | 1.0000 |
| Related SAEs | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| Death | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
Adverse events were coded according to MedDRA (version 24.1). Related AEs were those related to the study vaccine, referring to the adverse events as “possibly related,” “probably related,” or “definitely related” to the vaccine, which were identified by the principal investigators.
The incidence of each adverse reaction was similar in the 2 dose groups (50 and 100 μg), with the highest incidence of local adverse reactions (pain, swelling, erythra, and pruritus at the vaccination site). The highest incidence of local adverse reactions was pain, with an incidence of 61.11% and 67.78% in the 2 dose groups, respectively. The incidence of systemic adverse reactions in the 100-μg group was higher than the 50-μg group, and mainly included fever, weakness, headache, and fatigue. The highest incidence of systemic adverse reactions was fever, with an incidence of 8.89% and 21.67% in the 2 dose groups, respectively (Table 3).
Solicited adverse reactions (ARs) within 14 days after 1st or 2nd vaccinations, unsolicited adverse reactions within 28 days after each dose, graded by the National Medical Products Administration criteria.
| Adverse reactions - no. (%) | 50 μg group (N=180) | 100 μg group (N=180) | Placebo group (N=60) | P value |
|---|---|---|---|---|
| Solicited adverse reactions | ||||
| Any | 124 (68.89) | 134 (74.44) | 2 (3.33) | <0.0001 |
| ≥Grade 3 | 2 (1.11) | 12 (6.67) | 0 (0) | 0.0053 |
| Local adverse reactions | ||||
| Any | 120 (66.67) | 126 (70.00) | 0 (0) | <0.0001 |
| ≥Grade 3 | 2 (1.11) | 0 (0) | 0 (0) | 0.6318 |
| Pain | 110 (61.11) | 122 (67.78) | 0 (0) | <0.0001 |
| Swelling | 39 (21.67) | 40 (22.22) | 0 (0) | <0.0001 |
| Redness | 42 (23.33) | 36 (20.00) | 0 (0) | <0.0001 |
| Itch | 37 (20.56) | 34 (18.89) | 0 (0) | <0.0001 |
| Erythra | 3 (1.67) | 3 (1.67) | 0 (0) | <0.0001 |
| Induration | 1 (0.56) | 3 (1.67) | 0 (0) | <0.0001 |
| Systemic adverse reactions | ||||
| Any | 30 (16.67) | 57 (31.67) | 2 (3.33) | <0.0001 |
| ≥Grade 3 | 0 (0) | 12 (6.67) | 0 (0) | 0.0001 |
| Fever | 16 (8.89) | 39 (21.67) | 0 (0) | <0.0001 |
| Weakness | 16 (8.89) | 27 (15.00) | 1 (1.67) | 0.0065 |
| Headache | 14 (7.78) | 27 (15.00) | 1 (1.67) | 0.0036 |
| Fatigue | 12 (6.67) | 18 (10.00) | 0 (0) | 0.0166 |
| Anorexia | 5 (2.78) | 7 (3.89) | 0 (0) | 0.3757 |
| Nausea | 5 (2.78) | 7 (3.89) | 0 (0) | 0.3757 |
| Joint pain | 4 (2.22) | 9 (5.00) | 0 (0) | 0.1200 |
| Muscle pain (non-injection site) | 3 (1.67) | 8 (4.44) | 0 (0) | 0.1232 |
| Chills | 1 (0.56) | 4 (2.22) | 0 (0) | 0.3698 |
| Diarrhea | 2 (1.11) | 2 (1.11) | 0 (0) | 1.0000 |
| Chest pain | 0 (0) | 1 (0.56) | 1 (1.67) | 0.2656 |
| Unsolicited adverse reactions | ||||
| Any | 61 (33.89) | 88 (48.89) | 5 (8.33) | <0.0001 |
| ≥Grade 3 | 2 (1.11) | 0 (0) | 0 (0) | 0.6318 |
Adverse events were coded according to MedDRA (version 24.1). Adverse reactions shown were those related to the study vaccine, referring to the adverse events as “possibly related,” “probably related,” or “definitely related” to the vaccine, which were identified by the principal investigators.
Humoral immunity of LVRNA009 in the phase II clinical study
LVRNA009 induced high levels of neutralizing antibodies against the live SARS-CoV-2 (wild type) on days 14 and 28 after the full immunization, reaching a peak on day 14 with geometric mean titers (GMTs) of 995 and 1403 for 50 ug and 100 ug, respectively. The GMTs decreased on day 28, but remained as high as 615 and 763, respectively. The virus neutralizing antibodies in the 100-μg group were significantly higher than the 50-μg group on days 14 and 28 after full vaccination, suggesting that there may be a dose-dependent increase in the level of neutralizing antibodies against the live SARS-CoV-2 (wild type) after 2 doses of vaccination (Fig 5A).
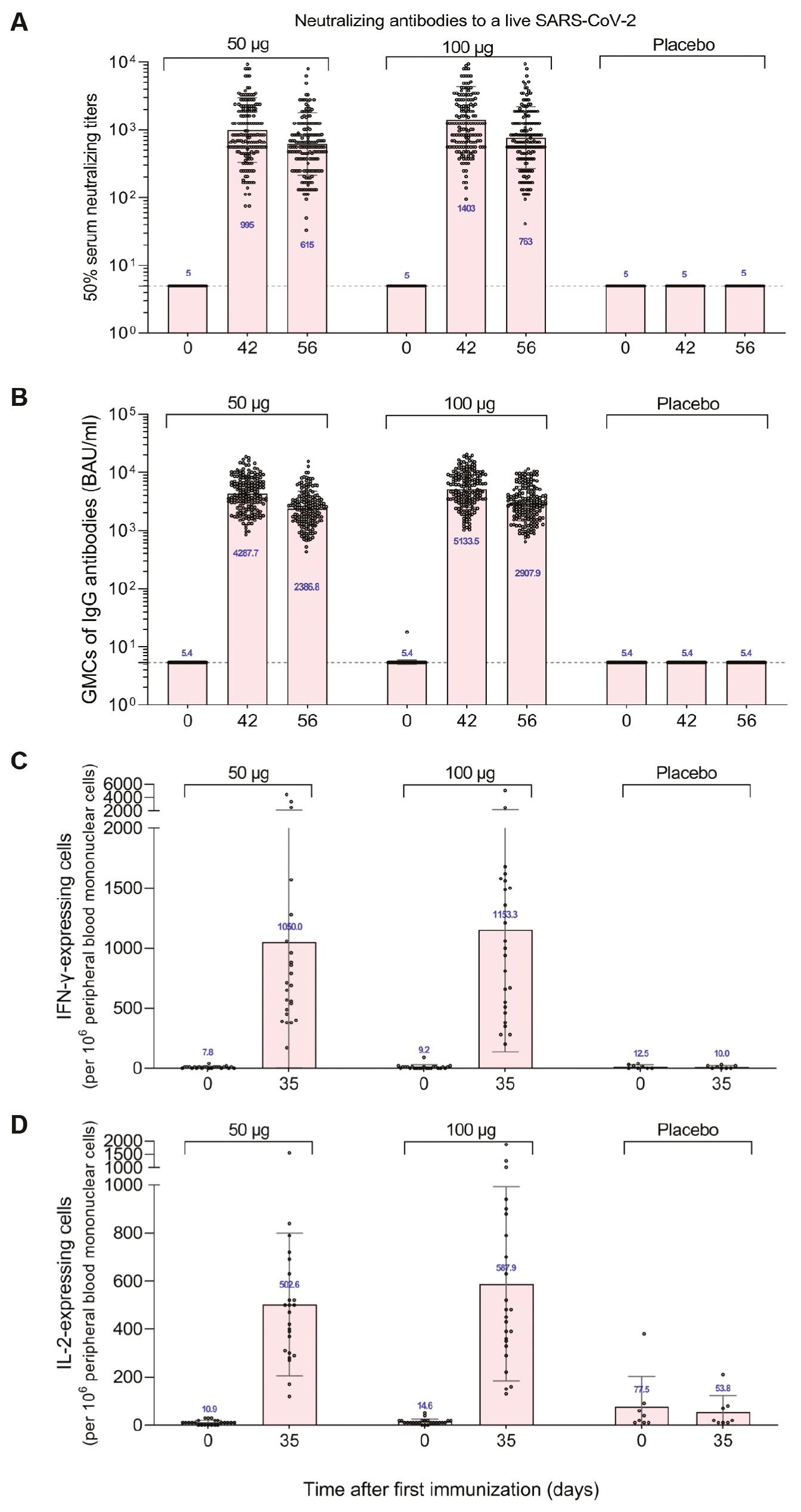
Humoral immunity effects and T cell responses induced by LVRNA009 vaccination. (A) Geometric mean titers of neutralizing antibodies to alive SARS-CoV-2 and (B) Geometric mean concentrations of the S-protein-specific IgG in serum samples of the participants collected at baseline (day 0), 14 days and 28 days after the full vaccination (day 42, day 56). (C) IFN-γ-expressing peripheral blood mononuclear cells and (D) IL-2-expressing peripheral blood mononuclear cells from blood samples of the participants collected at baseline (day 0) and 7 days after the full vaccination (day 35) by ELISPOT. Dot represents each sample. Bars represent standard deviation (SD). Numbers above dots show the geometric mean titre (A and B) or the mean value (C and D) of the group. Dashed line indicates the lower limit of quantification.
The geometric mean concentration (GMC) of IgG antibodies in the 50- and 100-μg groups peaked at 4287.7 binding antibody units (BAU)/ml and 5133.5 BAU/ml on days 14 and 28 after the full immunization, respectively, and decreased slightly on day 28, but remained high at 2386.80 BAU/ml and 2907.9 BAU/ml, respectively. There was no change in IgG antibody GMC on days 14 and 28 after full immunization in the placebo group compared to pre-immunization. The IgG antibody geometric mean concentration (GMC) and geometric mean growth multiple (GMI) 100-μg group were significantly higher than the 50-μg group on days 14 and 28 after full immunization (Fig 5B).
The positive conversion rate for WT strain live virus neutralizing and IgG antibodies were consistent at all time points after full immunization. The kappa coefficient was 1.00 and the agreement rate was 100%.
LVRNA009 cellular immunity in the phase II clinical study
The study groups induced significant cellular immune effects after two doses of vaccine. The number of cells expressing specific cytokines (IFN-γ, IL-2, IL-4, and IL-13) were significantly higher in the 50- and 100-μg test groups on day 7 after the full vaccination when compared to prior to the first vaccination, with the highest number of IFN-γ-expressing positive cells (1050/106 and 1153/106 cells, respectively). There was no change in the mean number of specific cytokine-expressing cells before and after placebo group vaccination (Fig 5C & 5D).
DISCUSSION
To control the global pandemic caused by SARS-CoV-2 it is necessary to know whether vaccines provide long-term protection against the current SARS-CoV-2 variants. In the current study an mRNA vaccine encoding the Wuhan-Hu-1 strain S protein was designed and developed. Immunization of mice with LVRNA009 and challenged with the Wuhan-Hu-1 strain inhibited RNA replication and alleviated pathologic changes in mouse lungs. In addition, LVRNA009 induced robust antigen-specific Th1 cell responses in mice by day 7 after full immunization and 8 days after virus infection, which is important for long-term prevention against the virus.
To evaluate the effectiveness of COVID-19 vaccines, the neutralization antibody titer (VNT) after vaccination is considered the main indicator [34–35]. High VNTs against Wuhan-Hu-1 strain and Omicron variants can be detected in mice after 8 months with LVRNA009 prime and boost immunizations. These mice were well protected against Omicron BA.1.1 infection and had a significantly reduced viral load in lung tissues. Moreover, when mice were boosted again with LVRNA009, the VNTs against Omicron BA.5 were significantly increased compared to the mice before the 2nd boost (Fig 3D).
During the development of vaccines against the COVID-19 variants, researchers have paid more attention to immunogenic imprinting, which is also known as original antigenic sin [36]. However, the concept is not very conclusive and represents positive and negative feedback [37]. A research group analyzed the effectiveness of cross-neutralization against pseudotyped BA.4/5 and BA.2.75 subvariants in two groups of individuals who had received inactivated vaccines. The results showed that the antibody response to subsequent antigenic stimulation was limited in individuals with BA.2 breakthrough infections, suggesting that the initial immune background has a role in shaping the immune response. The researchers concluded that original antigenic sin was observed in BA.1 and BA.2 breakthrough infections [36]. Another research group evaluated antibody responses in participants with COVID-19 monovalent mRNA vaccine of different immunization procedures or patients who had Omicron breakthrough infections after vaccination. Various analyses indicated that the original spike present in bivalent vaccines creates significant immunologic imprinting, hindering the development of antibodies against the BA.5 element and ultimately compromising the desired effectiveness. Therefore, it is advised to eliminate the original spike from upcoming COVID-19 vaccines to enhance the immune response to specific variants.
It is undeniable that most of the global population has been immunologically exposed to the original SARS-CoV-2 spike through vaccination, infection, or a combination of both. It is still worthwhile to evaluate the value of vaccine developed from the ancestral strain. In the current study the 2nd LVRNA009 boost induced much higher titers of neutralization antibodies against SARS-CoV-2 variants and protected the mice from Omicron variant challenge. The neutralization antibodies were assessed with live virus, not a pseudotyped virus assay, which reflects the real-world situation. Moreover, imprinting with pre-Omicron strains was shown to be uncompromised protection against severe disease [38].
The limitation of this study was that we only evaluated the long-term protection in mice. More convincing data should be collected from clinical studies in the future. The phase I and II LVRNA009 clinical studies have shown promising safety and immunogenicity profiles. A portion of the sera samples were tested for neutralizing antibody (nAb) activity against Omicron BA.1.1 on day 14 after the full immunization. Statistically increased nAb titers were detected in comparison with baseline but without immunization. These titers were not very strong and slightly different from the experimental results in mice (unpublished data). Another limitation for these studies is that the latest variants could not be evaluated due to a lack of the authentic viruses. We should notice that all of the recent virus strains still belong to Omicron sub-lineages and share most of the key S protein mutations with the variants that were tested. Thus, it is likely that the prevention potential of an LVRNA009 booster to the latest Omicron variants are also promising.
Combined with the encouraged immunogenicity results in mice with a 2nd LVRNA009 boost, it shall be helpful to use LVRNA009 as a boost vaccination for building anti-SARS-CoV-2 variants immunity in communities.