- Record: found
- Abstract: found
- Article: found
Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV
Read this article at
Abstract
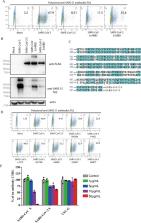
Since 2002, beta coronaviruses (CoV) have caused three zoonotic outbreaks, SARS-CoV in 2002–2003, MERS-CoV in 2012, and the newly emerged SARS-CoV-2 in late 2019. However, little is currently known about the biology of SARS-CoV-2. Here, using SARS-CoV-2 S protein pseudovirus system, we confirm that human angiotensin converting enzyme 2 (hACE2) is the receptor for SARS-CoV-2, find that SARS-CoV-2 enters 293/hACE2 cells mainly through endocytosis, that PIKfyve, TPC2, and cathepsin L are critical for entry, and that SARS-CoV-2 S protein is less stable than SARS-CoV S. Polyclonal anti-SARS S1 antibodies T62 inhibit entry of SARS-CoV S but not SARS-CoV-2 S pseudovirions. Further studies using recovered SARS and COVID-19 patients’ sera show limited cross-neutralization, suggesting that recovery from one infection might not protect against the other. Our results present potential targets for development of drugs and vaccines for SARS-CoV-2.
Abstract
SARS-CoV-2 has spread globally. Here, the authors characterize the entry pathway of SARS-CoV-2, show that the SARS-CoV-2 spike protein is less stable than that of SARS-CoV, and show limited cross-neutralization activities between SARS-CoV and SARS-CoV-2 sera.
Related collections
Most cited references12
- Record: found
- Abstract: found
- Article: not found
Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment.
- Record: found
- Abstract: found
- Article: not found
Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein.
- Record: found
- Abstract: found
- Article: not found