- Record: found
- Abstract: found
- Article: found
The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
Read this article at
Abstract
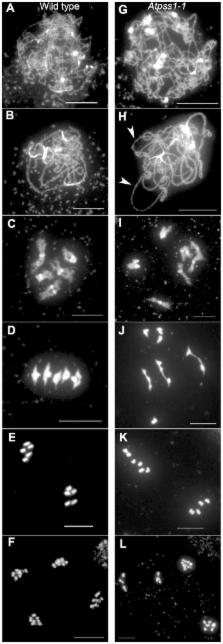
Meiotic crossovers (COs) shape genetic diversity by mixing homologous chromosomes at each generation. CO distribution is a highly regulated process. CO assurance forces the occurrence of at least one obligatory CO per chromosome pair, CO homeostasis smoothes out the number of COs when faced with variation in precursor number and CO interference keeps multiple COs away from each other along a chromosome. In several organisms, it has been shown that cytoskeleton forces are transduced to the meiotic nucleus via KASH- and SUN-domain proteins, to promote chromosome synapsis and recombination. Here we show that the Arabidopsis kinesin AtPSS1 plays a major role in chromosome synapsis and regulation of CO distribution. In Atpss1 meiotic cells, chromosome axes and DNA double strand breaks (DSBs) appear to form normally but only a variable portion of the genome synapses and is competent for CO formation. Some chromosomes fail to form the obligatory CO, while there is an increased CO density in competent regions. However, the total number of COs per cell is unaffected. We further show that the kinesin motor domain of AtPSS1 is required for its meiotic function, and that AtPSS1 interacts directly with WIP1 and WIP2, two KASH-domain proteins. Finally, meiocytes missing AtPSS1 and/or SUN proteins show similar meiotic defects suggesting that AtPSS1 and SUNs act in the same pathway. This suggests that forces produced by the AtPSS1 kinesin and transduced by WIPs/SUNs, are required to authorize complete synapsis and regulate maturation of recombination intermediates into COs. We suggest that a form of homeostasis applies, which maintains the total number of COs per cell even if only a part of the genome is competent for CO formation.
Author Summary
In species that reproduce sexually, diploid individuals have two copies of each chromosome, inherited from their father and mother. During a special cell division called meiosis, these two sets of chromosomes are mixed by homologous recombination to give genetically unique chromosomes that will be transmitted to the next generation. Homologous recombination processes are highly controlled in terms of number and localization of events within and among chromosomes. Disruption of this control (a lack of or improper positioning of homologous recombination events) causes deleterious chromosome associations in the offspring. Using the model plant Arabidopsis thaliana we reveal here that the AtPSS1 gene is required for proper localization of these homologous recombination events along the genome. We also show that AtPSS1, which belongs to a family of proteins able to move along the cytoskeleton, is likely part of a module that allows cytoplasmic forces to be transmitted through the nucleus envelope to promote chromosome movements during homologous recombination progression.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus.
- Record: found
- Abstract: found
- Article: not found
Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes.
- Record: found
- Abstract: found
- Article: not found