- Record: found
- Abstract: found
- Article: found
DUX4c Is Up-Regulated in FSHD. It Induces the MYF5 Protein and Human Myoblast Proliferation
Read this article at
Abstract
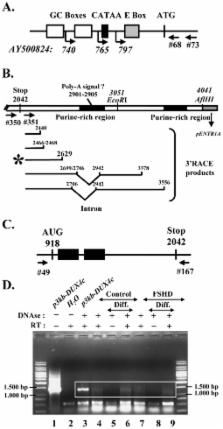
Facioscapulohumeral muscular dystrophy (FSHD) is a dominant disease linked to contractions of the D4Z4 repeat array in 4q35. We have previously identified a double homeobox gene ( DUX4) within each D4Z4 unit that encodes a transcription factor expressed in FSHD but not control myoblasts. DUX4 and its target genes contribute to the global dysregulation of gene expression observed in FSHD. We have now characterized the homologous DUX4c gene mapped 42 kb centromeric of the D4Z4 repeat array. It encodes a 47-kDa protein with a double homeodomain identical to DUX4 but divergent in the carboxyl-terminal region. DUX4c was detected in primary myoblast extracts by Western blot with a specific antiserum, and was induced upon differentiation. The protein was increased about 2-fold in FSHD versus control myotubes but reached 2-10-fold induction in FSHD muscle biopsies. We have shown by Western blot and by a DNA-binding assay that DUX4c over-expression induced the MYF5 myogenic regulator and its DNA-binding activity. DUX4c might stabilize the MYF5 protein as we detected their interaction by co-immunoprecipitation. In keeping with the known role of Myf5 in myoblast accumulation during mouse muscle regeneration DUX4c over-expression activated proliferation of human primary myoblasts and inhibited their differentiation. Altogether, these results suggested that DUX4c could be involved in muscle regeneration and that changes in its expression could contribute to the FSHD pathology.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy.
- Record: found
- Abstract: found
- Article: not found
DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1.
- Record: found
- Abstract: found
- Article: not found