- Record: found
- Abstract: found
- Article: found
Deep Learning Neural Networks Highly Predict Very Early Onset of Pluripotent Stem Cell Differentiation

Read this article at
Summary
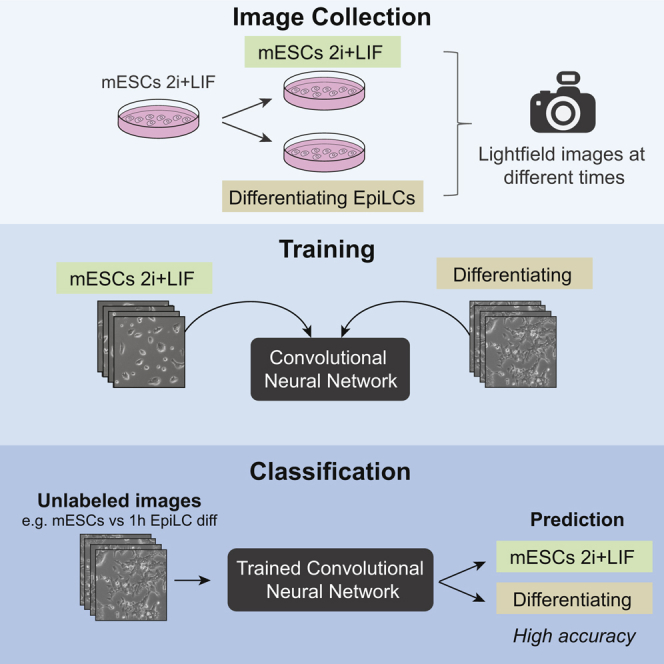
Deep learning is a significant step forward for developing autonomous tasks. One of its branches, computer vision, allows image recognition with high accuracy thanks to the use of convolutional neural networks (CNNs). Our goal was to train a CNN with transmitted light microscopy images to distinguish pluripotent stem cells from early differentiating cells. We induced differentiation of mouse embryonic stem cells to epiblast-like cells and took images at several time points from the initial stimulus. We found that the networks can be trained to recognize undifferentiated cells from differentiating cells with an accuracy higher than 99%. Successful prediction started just 20 min after the onset of differentiation. Furthermore, CNNs displayed great performance in several similar pluripotent stem cell (PSC) settings, including mesoderm differentiation in human induced PSCs. Accurate cellular morphology recognition in a simple microscopic set up may have a significant impact on how cell assays are performed in the near future.
Graphical Abstract
Highlights
-
•
Deep learning (DL) neural networks can efficiently classify light microscopy images
-
•
DL recognize PSC from 1 h differentiating epiblast-like cells
-
•
DL discriminates images of mouse ESCs in 2i + LIF from those grown in FBS + LIF
-
•
DL accurately classifies images of human iPSCs from those of early mesodermal cells
Abstract
In this article, Miriuka and colleagues show that deep learning convolutional neural networks can be trained to accurately classify light microscopy images of pluripotent stem cells from those of early differentiating cells, only minutes after the differentiation stimulus. These algorithms thus provide novel tools to quantitatively characterize subtle changes in cell morphology.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells.

- Record: found
- Abstract: found
- Article: not found