- Record: found
- Abstract: found
- Article: not found
Human Skin Fungal Diversity
Read this article at
Abstract
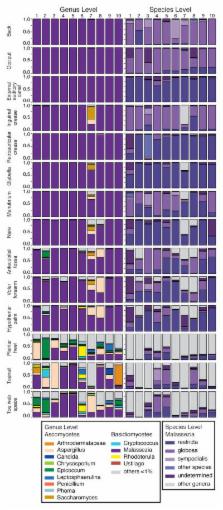
Traditional culture-based methods have incompletely defined the etiology of common recalcitrant human fungal skin diseases including athlete’s foot and toenail infections. Skin protects humans from invasion by pathogenic microorganisms, while providing a home for diverse commensal microbiota 1 . Bacterial genomic sequence data have generated novel hypotheses about species and community structures underlying human disorders 2, 3, 4 . However, microbial diversity is not limited to bacteria; microorganisms such as fungi also play major roles in microbial community stability, human health and disease 5 . Genomic methodologies to identify fungal species and communities have been limited compared with tools available for bacteria 6 . Fungal evolution can be reconstructed with phylogenetic markers, including ribosomal RNA gene regions and other highly conserved genes 7 . Here, we sequenced and analyzed fungal communities of 14 skin sites in 10 healthy adults. Eleven core body and arm sites were dominated by Malassezia fungi, with species-level classifications revealing greater topographical resolution between sites. By contrast, three foot sites, plantar heel, toenail, and toeweb, exhibited tremendous fungal diversity. Concurrent analysis of bacterial and fungal communities demonstrated that skin physiologic attributes and topography differentially shape these two microbial communities. These results provide a framework for future investigation of interactions between pathogenic and commensal fungal and bacterial communities in maintaining human health and contributing to disease pathogenesis.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis.
- Record: found
- Abstract: found
- Article: not found
