- Record: found
- Abstract: found
- Article: found
Macroalgal virosphere assists with host–microbiome equilibrium regulation and affects prokaryotes in surrounding marine environments
Read this article at
Abstract
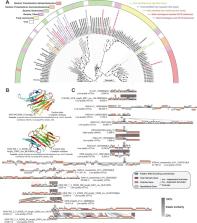
The microbiomes in macroalgal holobionts play vital roles in regulating macroalgal growth and ocean carbon cycling. However, the virospheres in macroalgal holobionts remain largely underexplored, representing a critical knowledge gap. Here we unveil that the holobiont of kelp ( Saccharina japonica) harbors highly specific and unique epiphytic/endophytic viral species, with novelty (99.7% unknown) surpassing even extreme marine habitats (e.g. deep-sea and hadal zones), indicating that macroalgal virospheres, despite being closest to us, are among the least understood. These viruses potentially maintain microbiome equilibrium critical for kelp health via lytic-lysogenic infections and the expression of folate biosynthesis genes. In-situ kelp mesocosm cultivation and metagenomic mining revealed that kelp holobiont profoundly reshaped surrounding seawater and sediment virus–prokaryote pairings through changing surrounding environmental conditions and virus–host migrations. Some kelp epiphytic viruses could even infect sediment autochthonous bacteria after deposition. Moreover, the presence of ample viral auxiliary metabolic genes for kelp polysaccharide (e.g. laminarin) degradation underscores the underappreciated viral metabolic influence on macroalgal carbon cycling. This study provides key insights into understanding the previously overlooked ecological significance of viruses within macroalgal holobionts and the macroalgae–prokaryotes–virus tripartite relationship.
Related collections
Most cited references93

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.

- Record: found
- Abstract: found
- Article: found