- Record: found
- Abstract: found
- Article: found
DNA-informed breeding of rosaceous crops: promises, progress and prospects
Read this article at
Abstract
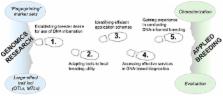
Crops of the Rosaceae family provide valuable contributions to rural economies and human health and enjoyment. Sustained solutions to production challenges and market demands can be met with genetically improved new cultivars. Traditional rosaceous crop breeding is expensive and time-consuming and would benefit from improvements in efficiency and accuracy. Use of DNA information is becoming conventional in rosaceous crop breeding, contributing to many decisions and operations, but only after past decades of solved challenges and generation of sufficient resources. Successes in deployment of DNA-based knowledge and tools have arisen when the ‘chasm’ between genomics discoveries and practical application is bridged systematically. Key steps are establishing breeder desire for use of DNA information, adapting tools to local breeding utility, identifying efficient application schemes, accessing effective services in DNA-based diagnostics and gaining experience in integrating DNA information into breeding operations and decisions. DNA-informed germplasm characterization for revealing identity and relatedness has benefitted many programs and provides a compelling entry point to reaping benefits of genomics research. DNA-informed germplasm evaluation for predicting trait performance has enabled effective reallocation of breeding resources when applied in pioneering programs. DNA-based diagnostics is now expanding from specific loci to genome-wide considerations. Realizing the full potential of this expansion will require improved accuracy of predictions, multi-trait DNA profiling capabilities, streamlined breeding information management systems, strategies that overcome plant-based features that limit breeding progress and widespread training of current and future breeding personnel and allied scientists.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Marker-assisted selection: an approach for precision plant breeding in the twenty-first century.
- Record: found
- Abstract: not found
- Article: not found
Molecular plant breeding as the foundation for 21st century crop improvement.
- Record: found
- Abstract: found
- Article: not found