- Record: found
- Abstract: found
- Article: found
Predicting continuous ground reaction forces from accelerometers during uphill and downhill running: a recurrent neural network solution
Read this article at
Abstract
Background
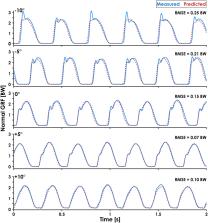
Ground reaction forces (GRFs) are important for understanding human movement, but their measurement is generally limited to a laboratory environment. Previous studies have used neural networks to predict GRF waveforms during running from wearable device data, but these predictions are limited to the stance phase of level-ground running. A method of predicting the normal (perpendicular to running surface) GRF waveform using wearable devices across a range of running speeds and slopes could allow researchers and clinicians to predict kinetic and kinematic variables outside the laboratory environment.
Purpose
We sought to develop a recurrent neural network capable of predicting continuous normal (perpendicular to surface) GRFs across a range of running speeds and slopes from accelerometer data.
Methods
Nineteen subjects ran on a force-measuring treadmill at five slopes (0°, ±5°, ±10°) and three speeds (2.5, 3.33, 4.17 m/s) per slope with sacral- and shoe-mounted accelerometers. We then trained a recurrent neural network to predict normal GRF waveforms frame-by-frame. The predicted versus measured GRF waveforms had an average ± SD RMSE of 0.16 ± 0.04 BW and relative RMSE of 6.4 ± 1.5% across all conditions and subjects.
Results
The recurrent neural network predicted continuous normal GRF waveforms across a range of running speeds and slopes with greater accuracy than neural networks implemented in previous studies. This approach may facilitate predictions of biomechanical variables outside the laboratory in near real-time and improves the accuracy of quantifying and monitoring external forces experienced by the body when running.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
Long Short-Term Memory

- Record: found
- Abstract: found
- Article: found
SciPy 1.0: fundamental algorithms for scientific computing in Python
