- Record: found
- Abstract: found
- Article: found
Comprehensive structure and functional adaptations of the yeast nuclear pore complex
Read this article at
SUMMARY
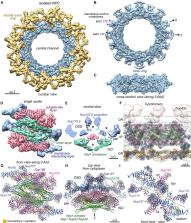
Nuclear pore complexes (NPCs) mediate the nucleocytoplasmic transport of macromolecules. Here we provide a structure of the isolated yeast NPC in which the inner ring is resolved by cryo-EM at sub-nanometer resolution to show how flexible connectors tie together different structural and functional layers. These connectors may be targets for phosphorylation and regulated disassembly in cells with an open mitosis. Moreover, some nucleoporin pairs and transport factors have similar interaction motifs, which suggests an evolutionary and mechanistic link between assembly and transport. We provide evidence for three major NPC variants that may foreshadow functional specializations at the nuclear periphery. Cryo-electron tomography extended these studies, providing a model of the in situ NPC with a radially expanded inner ring. Our comprehensive model reveals features of the nuclear basket and central transporter, suggests a role for the lumenal Pom152 ring in restricting dilation, and highlights structural plasticity that may be required for transport.
In brief
A comprehensive model of the yeast NPC reveals an interconnected architecture of the core scaffold and provides an understanding of the isoforms and structural plasticity that may be associated with different functional states.
Graphical Abstract

Related collections
Most cited references115

- Record: found
- Abstract: found
- Article: found
Highly accurate protein structure prediction with AlphaFold
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: found
- Article: not found