- Record: found
- Abstract: found
- Article: found
DNA Fingerprinting of Mycobacterium leprae Strains Using Variable Number Tandem Repeat (VNTR) - Fragment Length Analysis (FLA)
Read this article at
Abstract
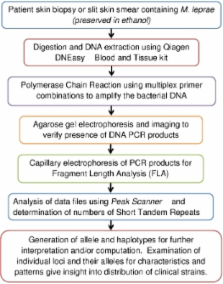
The study of the transmission of leprosy is particularly difficult since the causative agent, Mycobacterium leprae, cannot be cultured in the laboratory. The only sources of the bacteria are leprosy patients, and experimentally infected armadillos and nude mice. Thus, many of the methods used in modern epidemiology are not available for the study of leprosy. Despite an extensive global drug treatment program for leprosy implemented by the WHO 1, leprosy remains endemic in many countries with approximately 250,000 new cases each year. 2 The entire M. leprae genome has been mapped 3,4 and many loci have been identified that have repeated segments of 2 or more base pairs (called micro- and minisatellites). 5 Clinical strains of M. leprae may vary in the number of tandem repeated segments (short tandem repeats, STR) at many of these loci. 5,6,7 Variable number tandem repeat (VNTR) 5 analysis has been used to distinguish different strains of the leprosy bacilli. Some of the loci appear to be more stable than others, showing less variation in repeat numbers, while others seem to change more rapidly, sometimes in the same patient. While the variability of certain VNTRs has brought up questions regarding their suitability for strain typing 7,8,9, the emerging data suggest that analyzing multiple loci, which are diverse in their stability, can be used as a valuable epidemiological tool. Multiple locus VNTR analysis (MLVA) 10 has been used to study leprosy evolution and transmission in several countries including China 11,12, Malawi 8, the Philippines 10,13, and Brazil 14. MLVA involves multiple steps. First, bacterial DNA is extracted along with host tissue DNA from clinical biopsies or slit skin smears (SSS). 10 The desired loci are then amplified from the extracted DNA via polymerase chain reaction (PCR). Fluorescently-labeled primers for 4-5 different loci are used per reaction, with 18 loci being amplified in a total of four reactions. 10 The PCR products may be subjected to agarose gel electrophoresis to verify the presence of the desired DNA segments, and then submitted for fluorescent fragment length analysis (FLA) using capillary electrophoresis. DNA from armadillo passaged bacteria with a known number of repeat copies for each locus is used as a positive control. The FLA chromatograms are then examined using Peak Scanner software and fragment length is converted to number of VNTR copies (allele). Finally, the VNTR haplotypes are analyzed for patterns, and when combined with patient clinical data can be used to track distribution of strain types.
Related collections
Most cited references12
- Record: found
- Abstract: not found
- Article: not found
Chemotherapy of leprosy for control programmes.
- Record: found
- Abstract: found
- Article: not found
Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae.
- Record: found
- Abstract: found
- Article: not found