- Record: found
- Abstract: found
- Article: found
Base-resolution methylomes of gliomas bearing histone H3.3 mutations reveal a G34 mutant-specific signature shared with bone tumors
Read this article at
Abstract
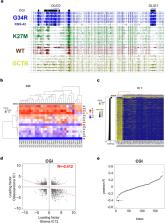
Two recurrent mutations, K27M and G34R/V, in H3F3A, encoding non-canonical histone H3.3, are reported in pediatric and young adult gliomas, whereas G34W mutation is prevalent in bone tumors. In contrast to K27M mutation, it remains elusive how G34 mutations affect the epigenome. Here we performed whole-genome bisulfite sequencing of four G34R-mutated gliomas and the G34V-mutated glioma cell line KNS-42 for comparison with gliomas harboring K27M and no mutations in H3F3A and with G34W-mutated bone tumors. G34R-mutated gliomas exhibited lower global methylation levels, similar CpG island (CGI) methylation levels, and compromised hypermethylation of telomere-proximal CGIs, compared to the other two glioma subgroups. Hypermethylated regions specific to G34R-mutated gliomas were enriched for CGIs, including those of OLIG1, OLIG2, and canonical histone genes in the HIST1 cluster. They were notably hypermethylated in osteosarcomas with, but not without, G34W mutation. Independent component analysis revealed that G34 mutation-specific components shared a significant similarity between glioma and osteosarcoma, suggesting that G34 mutations exert characteristic methylomic effects regardless of the tumor tissue-of-origin. CRISPR/Cas9-mediated disruption of G34V-allele in KNS-42 cells led to demethylation of a subset of CGIs hypermethylated in G34R-mutated gliomas. These findings will provide a basis for elucidating epigenomic roles of G34 oncohistone in tumorigenesis.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
DNA Methylation in Cancer and Aging.
- Record: found
- Abstract: found
- Article: not found
EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas

- Record: found
- Abstract: found
- Article: found