- Record: found
- Abstract: found
- Article: found
Complete human day 14 post-implantation embryo models from naive ES cells
Read this article at
Abstract
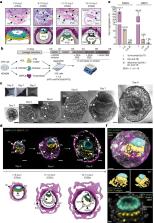
The ability to study human post-implantation development remains limited owing to ethical and technical challenges associated with intrauterine development after implantation 1 . Embryo-like models with spatially organized morphogenesis and structure of all defining embryonic and extra-embryonic tissues of the post-implantation human conceptus (that is, the embryonic disc, the bilaminar disc, the yolk sac, the chorionic sac and the surrounding trophoblast layer) remain lacking 1, 2 . Mouse naive embryonic stem cells have recently been shown to give rise to embryonic and extra-embryonic stem cells capable of self-assembling into post-gastrulation structured stem-cell-based embryo models with spatially organized morphogenesis (called SEMs) 3 . Here we extend those findings to humans using only genetically unmodified human naive embryonic stem cells (cultured in human enhanced naive stem cell medium conditions) 4 . Such human fully integrated and complete SEMs recapitulate the organization of nearly all known lineages and compartments of post-implantation human embryos, including the epiblast, the hypoblast, the extra-embryonic mesoderm and the trophoblast layer surrounding the latter compartments. These human complete SEMs demonstrated developmental growth dynamics that resemble key hallmarks of post-implantation stage embryogenesis up to 13–14 days after fertilization (Carnegie stage 6a). These include embryonic disc and bilaminar disc formation, epiblast lumenogenesis, polarized amniogenesis, anterior–posterior symmetry breaking, primordial germ-cell specification, polarized yolk sac with visceral and parietal endoderm formation, extra-embryonic mesoderm expansion that defines a chorionic cavity and a connecting stalk, and a trophoblast-surrounding compartment demonstrating syncytium and lacunae formation. This SEM platform will probably enable the experimental investigation of previously inaccessible windows of human early post implantation up to peri-gastrulation development.
Abstract
The culture of genetically unmodified human naive embryonic stem cells in specific growth conditions gives rise to structures that recapitulate those of post-implantation human embryos up to 13–14 days after fertilization.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data
- Record: found
- Abstract: found
- Article: not found
Integrating single-cell transcriptomic data across different conditions, technologies, and species
- Record: found
- Abstract: found
- Article: not found