- Record: found
- Abstract: found
- Article: found
Genomic Epidemiology of Major Extraintestinal Pathogenic Escherichia coli Lineages Causing Urinary Tract Infections in Young Women Across Canada
Read this article at
Abstract
Background
A few extraintestinal pathogenic Escherichia coli (ExPEC) multilocus sequence types (STs) cause the majority of community-acquired urinary tract infections (UTIs). We examine the genomic epidemiology of major ExPEC lineages, specifically factors associated with intestinal acquisition.
Methods
A total of 385 women with UTI caused by E. coli across Canada were asked about their diet, travel, and other exposures. Genome sequencing was used to determine both ST and genomic similarity. Logistic regression was used to identify factors associated with the acquisition of and infection with major ExPEC STs relative to minor ExPEC STs.
Results
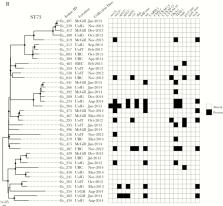
ST131, ST69, ST73, ST127, and ST95 were responsible for 54% of all UTIs. Seven UTI clusters were identified, but genomes from the ST95, ST127, and ST420 clusters exhibited as few as 3 single nucleotide variations across the entire genome, suggesting recent acquisition. Furthermore, we identified a cluster of UTIs caused by 6 genetically-related ST1193 isolates carrying mutations in gyrA and parC. The acquisition of and infection with ST69, ST95, ST127, and ST131 were all associated with increased travel. The consumption of high-risk foods such as raw meat or vegetables, undercooked eggs, and seafood was associated with acquisition of and infection with ST69, ST127, and ST131, respectively.
Related collections
Most cited references22
- Record: found
- Abstract: not found
- Article: not found
Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM
- Record: found
- Abstract: found
- Article: not found
Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance
- Record: found
- Abstract: found
- Article: not found