- Record: found
- Abstract: found
- Article: found
Integrating genomics for chickpea improvement: achievements and opportunities
Read this article at
Abstract
Key message
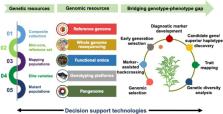
Integration of genomic technologies with breeding efforts have been used in recent years for chickpea improvement. Modern breeding along with low cost genotyping platforms have potential to further accelerate chickpea improvement efforts.
Abstract
The implementation of novel breeding technologies is expected to contribute substantial improvements in crop productivity. While conventional breeding methods have led to development of more than 200 improved chickpea varieties in the past, still there is ample scope to increase productivity. It is predicted that integration of modern genomic resources with conventional breeding efforts will help in the delivery of climate-resilient chickpea varieties in comparatively less time. Recent advances in genomics tools and technologies have facilitated the generation of large-scale sequencing and genotyping data sets in chickpea. Combined analysis of high-resolution phenotypic and genetic data is paving the way for identifying genes and biological pathways associated with breeding-related traits. Genomics technologies have been used to develop diagnostic markers for use in marker-assisted backcrossing programmes, which have yielded several molecular breeding products in chickpea. We anticipate that a sequence-based holistic breeding approach, including the integration of functional omics, parental selection, forward breeding and genome-wide selection, will bring a paradigm shift in development of superior chickpea varieties. There is a need to integrate the knowledge generated by modern genomics technologies with molecular breeding efforts to bridge the genome-to-phenome gap. Here, we review recent advances that have led to new possibilities for developing and screening breeding populations, and provide strategies for enhancing the selection efficiency and accelerating the rate of genetic gain in chickpea.
Related collections
Most cited references72
- Record: found
- Abstract: found
- Article: not found
GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein.
- Record: found
- Abstract: found
- Article: not found
The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism.
- Record: found
- Abstract: found
- Article: not found