- Record: found
- Abstract: found
- Article: found
Profiling several key milk miRNAs and analysing their signalling pathways in dairy sheep breeds during peak and late lactation

Read this article at
Abstract
Background
The comprehensive understanding of microRNAs (miRNAs) in sheep milk during various lactation periods and their impact on milk yield and composition remains limited.
Objectives
This study aimed to investigate the expression patterns of four highly expressed miRNAs in sheep milk and their association with milk composition and yield parameters during peak and late lactation stages.
Methods
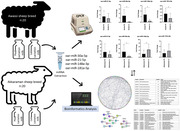
A total of 40 healthy 4‐year‐old Akkaraman ( n = 20) and Awassi ( n = 20) ewes registered with the Ministry of Agriculture and Forestry of the Republic of Türkiye were used in the present study. For miRNA isolation from milk, the Qiagen miRNeasy Serum/Plasma Advanced Kit was utilised following the manufacturer's instructions. The expression levels of miRNAs were assessed using Qiagen miRNA PCR Assays.
Results
The significant fold changes in the expression levels of oar‐miR‐30a‐5p, oar‐miR‐148a and oar‐miR‐181a were observed between peak and late lactation periods in the Awassi sheep breed. Conversely, only oar‐miR‐30a‐5p and oar‐miR‐148a exhibited statistically significant changes in the Akkaraman sheep breed during the same lactation periods. Furthermore, oar‐miR‐21‐5p demonstrated a significant fold change exclusively in peak lactation compared to Akkaraman and Awassi ewes.
Conclusions
The findings suggest that the expression of the analysed miRNAs is influenced by both the lactation stage and different sheep breeds. This study offers valuable insights into the relationship between key miRNA expressions in sheep milk and milk composition and yield parameters during peak and late lactation, contributing to the existing knowledge in this field.
Abstract
-
This study aimed to investigate the expression patterns of four highly expressed miRNAs in sheep milk and their association with milk composition and yield parameters during peak and late lactation stages.
-
Notably, significant fold changes in the expression levels of oar‐miR‐30a‐5p, oar‐miR‐148a, and oar‐miR‐181a were observed between peak and late lactation periods in the Awassi sheep breed.
-
oar‐miR‐21‐5p demonstrated a significant fold change exclusively in peak lactation compared to Akkaraman and Awassi ewes.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.

- Record: found
- Abstract: found
- Article: found
Metascape provides a biologist-oriented resource for the analysis of systems-level datasets

- Record: found
- Abstract: found
- Article: found