- Record: found
- Abstract: found
- Article: found
Toward a Predictive Understanding of Earth’s Microbiomes to Address 21st Century Challenges
Read this article at
ABSTRACT
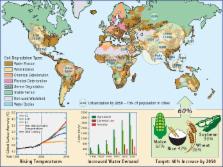
Microorganisms have shaped our planet and its inhabitants for over 3.5 billion years. Humankind has had a profound influence on the biosphere, manifested as global climate and land use changes, and extensive urbanization in response to a growing population. The challenges we face to supply food, energy, and clean water while maintaining and improving the health of our population and ecosystems are significant. Given the extensive influence of microorganisms across our biosphere, we propose that a coordinated, cross-disciplinary effort is required to understand, predict, and harness microbiome function. From the parallelization of gene function testing to precision manipulation of genes, communities, and model ecosystems and development of novel analytical and simulation approaches, we outline strategies to move microbiome research into an era of causality. These efforts will improve prediction of ecosystem response and enable the development of new, responsible, microbiome-based solutions to significant challenges of our time.
Related collections
Most cited references112
- Record: found
- Abstract: found
- Article: not found
Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013.
- Record: found
- Abstract: found
- Article: not found
Multiorganismal insects: diversity and function of resident microorganisms.
- Record: found
- Abstract: found
- Article: not found