- Record: found
- Abstract: found
- Article: found
Functionally-explicit sampling can answer key questions about the specificity of plant–microbe interactions
Read this article at
Abstract
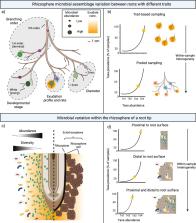
The rhizosphere is a nexus for plant–microbe interactions and, as a host-structured environment, a location of high activity for distinct microbes and plant species. Although our insights into this habitat have exploded in recent years, we are still limited in our ability to answer key questions about the specificity of these root-microbial relationships. In particular, it can be difficult to confirm or reject microbiome heritability in many plant systems and to pinpoint which microbial taxa are key to plant functioning. Like other host-structured environments, the rhizosphere is structurally, chemically, and biologically complex, driven largely by differences in root anatomy, location, and function. In this Correspondence, we describe a review of 377 “rhizosphere microbiome” research papers and demonstrate how matching a sampling method to the biological question can advance our understanding of host-microbe interactions in a functionally heterogeneous environment. We found that the vast majority of studies (92%) pool all roots from a root system during sampling, ignoring variation in microbial composition between roots of different function and limiting insight into key root-microbial relationships. Furthermore, approaches for removing root-associated microbes are highly variable and non-standard, complicating multi-study analyses. Our understanding of the strength and nature of host-microbe relationships in heterogenous host-microbiome environments can be clarified by targeting sampling to locations of high interaction. While the high complexity of the rhizosphere creates logistical challenges, we suggest that unambiguous language and refined approaches will improve our ability to match methods to research questions and advance our understanding of the specificity of plant-microbial interactions.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Plant–microbiome interactions: from community assembly to plant health
- Record: found
- Abstract: found
- Article: not found
Structure, variation, and assembly of the root-associated microbiomes of rice.
- Record: found
- Abstract: found
- Article: not found