- Record: found
- Abstract: found
- Article: found
Attenuation of Pseudomonas aeruginosa biofilm formation by Vitexin: A combinatorial study with azithromycin and gentamicin
Read this article at
Abstract
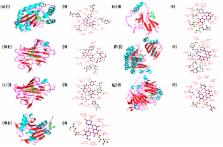
Microbial biofilm are communities of surface-adhered cells enclosed in a matrix of extracellular polymeric substances. Extensive use of antibiotics to treat biofilm associated infections has led to the emergence of multiple drug resistant strains. Pseudomonas aeruginosa is recognised as a model biofilm forming pathogenic bacterium. Vitexin, a polyphenolic group of phytochemical with antimicrobial property, has been studied for its antibiofilm potential against Pseudomonas aeruginosa in combination with azithromycin and gentamicin. Vitexin shows minimum inhibitory concentration (MIC) at 260 μg/ml. It’s antibiofilm activity was evaluated by safranin staining, protein extraction, microscopy methods, quantification of EPS and in vivo models using several sub-MIC doses. Various quorum sensing (QS) mediated phenomenon such as swarming motility, azocasein degrading protease activity, pyoverdin and pyocyanin production, LasA and LasB activity of the bacteria were also evaluated. Results showed marked attenuation in biofilm formation and QS mediated phenotype of Pseudomonas aeruginosa in presence of 110 μg/ml vitexin in combination with azithromycin and gentamicin separately. Molecular docking of vitexin with QS associated LuxR, LasA, LasI and motility related proteins showed high and reasonable binding affinity respectively. The study explores the antibiofilm potential of vitexin against P. aeruginosa which can be used as a new antibiofilm agent against microbial biofilm associated pathogenesis.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions.
- Record: found
- Abstract: found
- Article: not found