- Record: found
- Abstract: found
- Article: found
Identification of methylated genes and miRNA signatures in nasopharyngeal carcinoma by bioinformatics analysis
Read this article at
Abstract
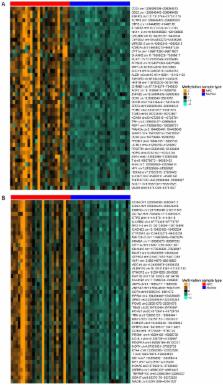
Nasopharyngeal carcinoma (NPC) is prevalent in several regions, including. Southern China and Southeast Asia, with high mortality. The present study aimed to explore the epigenetic mechanisms of NPC and to provide novel biomarkers for prognosis. Two methylation data sets (GSE52068 and GSE62336) were downloaded from the Gene Expression Omnibus database. Following pretreatment of the raw data, differentially methylated regions (DMRs) and differentially methylated CpG islands (DMCs) were identified between the NPC samples and normal tissue controls using COHCAP software. The overlapped DMRs and DMCs in the two data sets were extracted and associated to relevant genes. Enrichment analysis and protein-protein interaction (PPI) network analyses were performed on the identified genes using Database for Annotation, Visualization and Integration Discovery and Cytoscape, respectively. MicroRNAs (miRNAs) targeting the overlapped genes were identified based on the miRWalk database. NPC-related genes were analyzed with the Comparative Toxicogenomics Database. Multiple overlapping DMRs between the two data sets were identified and were associated with 1,854 hypermethylated and 18 hypomethylated genes, which were revealed to be enriched in certain pathways, including the mitogen-activated protein kinase (MAPK) signaling pathway and the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway. Several nodes in the predicted PPI network were highlighted, including proto-oncogene tyrosine-protein kinase SRC, SMAD family member 3 (SMAD3), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein ζ (YWHAZ) and Heat shock protein family A member 4 (HSPA4), all of which were hypomethylated. A total of 14 miRNAs were identified that correlated with the overlapped genes such as miRNA (miR)-148a-3p, which was predicted to target of HSPA4; and 17 genes were identified as related to NPC, including SMAD3 and SRC. miR129-2 was hypermethylated. Several novel methylated genes or miRNAs were suggested as biomarkers for NPC prognosis: Hypomethylation of SRC, SMAD3, YWHAZ and HSPA4, and hypermethylation of miR129-2 may be linked to poor prognosis of NPC.
Related collections
Most cited references26

- Record: found
- Abstract: found
- Article: not found
Epigenetic regulation of microRNA expression in colorectal cancer.

- Record: found
- Abstract: found
- Article: found