- Record: found
- Abstract: found
- Article: found
Antimicrobial resistance expansion in pathogens: a review of current mitigation strategies and advances towards innovative therapy
Read this article at
Abstract
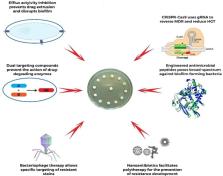
The escalating problem of antimicrobial resistance (AMR) proliferation in clinically important pathogens has become one of the biggest threats to human health and the global economy. Previous studies have estimated AMR-associated deaths and disability-adjusted life-years (DALYs) in many countries with a view to presenting a clearer picture of the global burden of AMR-related diseases. Recently, several novel strategies have been advanced to combat resistance spread. These include efflux activity inhibition, closing of mutant selection window (MSW), biofilm disruption, lytic bacteriophage particles, nanoantibiotics, engineered antimicrobial peptides, and the CRISPR-Cas9 gene-editing technique. The single or integrated deployment of these strategies has shown potentialities towards mitigating resistance and contributing to valuable therapeutic outcomes. Correspondingly, the new paradigm of personalized medicine demands innovative interventions such as improved and accurate point-of-care diagnosis and treatment to curtail AMR. The CRISPR-Cas system is a novel and highly promising nucleic acid detection and manipulating technology with the potential for application in the control of AMR. This review thus considers the specifics of some of the AMR-mitigating strategies, while noting their drawbacks, and discusses the advances in the CRISPR-based technology as an important point-of-care tool for tracking and curbing AMR in our fight against a looming ‘post-antibiotic’ era.
Related collections
Most cited references122

- Record: found
- Abstract: found
- Article: found
The antimicrobial activity of nanoparticles: present situation and prospects for the future
- Record: found
- Abstract: found
- Article: found