- Record: found
- Abstract: found
- Article: not found
Immature and Mature Human Astrovirus: Structure, Conformational Changes, and Similarities to Hepatitis E Virus ☆

Abstract
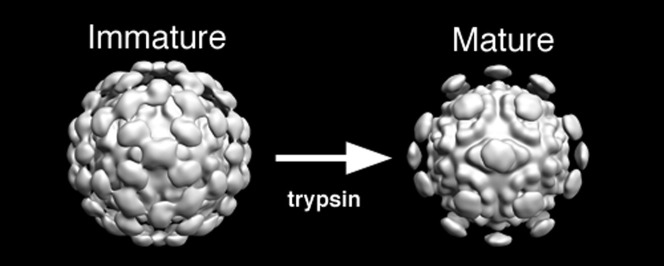
Human astroviruses (HAstVs) are a major cause of gastroenteritis. HAstV assembles from the structural protein VP90 and undergoes a cascade of proteolytic cleavages. Cleavage to VP70 is required for release of immature particles from cells, and subsequent cleavage by trypsin confers infectivity. We used electron cryomicroscopy and icosahedral image analysis to determine the first experimentally derived, three-dimensional structures of an immature VP70 virion and a fully proteolyzed, infectious virion. Both particles display T = 3 icosahedral symmetry and nearly identical solid capsid shells with diameters of ~ 350 Å. Globular spikes emanate from the capsid surface, yielding an overall diameter of ~ 440 Å. While the immature particles display 90 dimeric spikes, the mature capsid only displays 30 spikes, located on the icosahedral 2-fold axes. Loss of the 60 peripentonal spikes likely plays an important role in viral infectivity. In addition, immature HAstV bears a striking resemblance to the structure of hepatitis E virus (HEV)-like particles, as previously predicted from structural similarity of the crystal structure of the astrovirus spike domain with the HEV P-domain [Dong, J., Dong, L., Méndez, E. & Tao, Y. (2011). Crystal structure of the human astrovirus capsid spike. Proc. Natl. Acad. Sci. USA 108, 12681–12686]. Similarities between their capsid shells and dimeric spikes and between the sequences of their capsid proteins suggest that these viral families are phylogenetically related and may share common assembly and activation mechanisms.
Graphical Abstract
Related collections
Most cited references25

- Record: found
- Abstract: found
- Article: found
VIPERdb2: an enhanced and web API enabled relational database for structural virology
- Record: found
- Abstract: found
- Article: not found
Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity
- Record: found
- Abstract: not found
- Article: not found