- Record: found
- Abstract: found
- Article: found
Soil fertility relates to fungal-mediated decomposition and organic matter turnover in a temperate mountain forest
Read this article at
Summary
-
Fungi are known to exert a significant influence over soil organic matter (SOM) turnover, however understanding of the effects of fungal community structure on SOM dynamics and its consequences for ecosystem fertility is fragmentary.
-
Here we studied soil fungal guilds and SOM decomposition processes along a fertility gradient in a temperate mountain beech forest. High-throughput sequencing was used to investigate fungal communities. Carbon and nitrogen stocks, enzymatic activity and microbial respiration were measured.
-
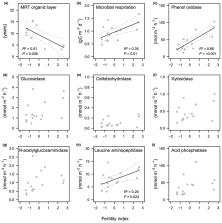
While ectomycorrhizal fungal abundance was not related to fertility, saprotrophic ascomycetes showed higher relative abundances under more fertile conditions. The activity of oxidising enzymes and respiration rates in mineral soil were related positively to fertility and saprotrophic fungi. In addition, organic layer carbon and nitrogen stocks were lower on the more fertile plots, although tree biomass and litter input were higher. Together, the results indicated a faster SOM turnover at the fertile end of the gradient.
-
We suggest that there is a positive feedback mechanism between SOM turnover and fertility that is mediated by soil fungi to a significant extent. By underlining the importance of fungi for soil fertility and plant growth, these findings furthermore emphasise the dependency of carbon cycling on fungal communities below ground.
Related collections
Most cited references82
- Record: found
- Abstract: not found
- Book Chapter: not found
AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS

- Record: found
- Abstract: found
- Article: found
VSEARCH: a versatile open source tool for metagenomics
- Record: found
- Abstract: found
- Article: not found