- Record: found
- Abstract: found
- Article: found
Size distribution and relationship of airborne SARS-CoV-2 RNA to indoor aerosol in hospital ward environments
Read this article at
Abstract
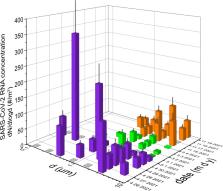
Aerosol particles proved to play a key role in airborne transmission of SARS-CoV-2 viruses. Therefore, their size-fractionated collection and analysis is invaluable. However, aerosol sampling in COVID departments is not straightforward, especially in the sub-500-nm size range. In this study, particle number concentrations were measured with high temporal resolution using an optical particle counter, and several 8 h daytime sample sets were collected simultaneously on gelatin filters with cascade impactors in two different hospital wards during both alpha and delta variants of concern periods. Due to the large number (152) of size-fractionated samples, SARS-CoV-2 RNA copies could be statistically analyzed over a wide range of aerosol particle diameters (70–10 µm). Our results revealed that SARS-CoV-2 RNA is most likely to exist in particles with 0.5–4 µm aerodynamic diameter, but also in ultrafine particles. Correlation analysis of particulate matter (PM) and RNA copies highlighted the importance of indoor medical activity. It was found that the daily maximum increment of PM mass concentration correlated the most with the number concentration of SARS-CoV-2 RNA in the corresponding size fractions. Our results suggest that particle resuspension from surrounding surfaces is an important source of SARS-CoV-2 RNA present in the air of hospital rooms.
Related collections
Most cited references69
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019
- Record: found
- Abstract: found
- Article: not found
A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster

- Record: found
- Abstract: found
- Article: found