- Record: found
- Abstract: found
- Article: found
Inducible expression of large gRNA arrays for multiplexed CRISPRai applications
Read this article at
Abstract
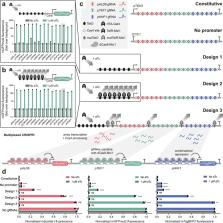
CRISPR gene activation and inhibition (CRISPRai) has become a powerful synthetic tool for influencing the expression of native genes for foundational studies, cellular reprograming, and metabolic engineering. Here we develop a method for near leak-free, inducible expression of a polycistronic array containing up to 24 gRNAs from two orthogonal CRISPR/Cas systems to increase CRISPRai multiplexing capacity and target gene flexibility. To achieve strong inducibility, we create a technology to silence gRNA expression within the array in the absence of the inducer, since we found that long gRNA arrays for CRISPRai can express themselves even without promoter. Using this method, we create a highly tuned and easy-to-use CRISPRai toolkit in the industrially relevant yeast, Saccharomyces cerevisiae, establishing the first system to combine simultaneous activation and repression, large multiplexing capacity, and inducibility. We demonstrate this toolkit by targeting 11 genes in central metabolism in a single transformation, achieving a 45-fold increase in succinic acid, which could be precisely controlled in an inducible manner. Our method offers a highly effective way to regulate genes and rewire metabolism in yeast, with principles of gRNA array construction and inducibility that should extend to other chassis organisms.
Abstract
CRISPR gene activation and inhibition has become a powerful synthetic tool for influencing the expression of native genes for foundational studies, cellular reprograming, and metabolic engineering. Here the authors demonstrate near leak-free, inducible expression of a polycistronic array containing up to 24 gRNAs from two orthogonal CRISPR/Cas systems.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression.
- Record: found
- Abstract: found
- Article: not found
CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes.
- Record: found
- Abstract: found
- Article: not found