- Record: found
- Abstract: found
- Article: found
Oncometabolite lactate enhances breast cancer progression by orchestrating histone lactylation-dependent c-Myc expression
Read this article at
Highlights
Abstract
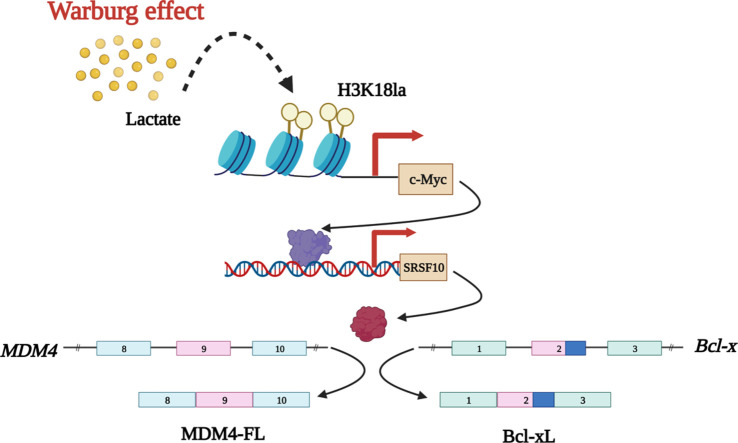
Due to the enhanced glycolytic rate, cancer cells generate lactate copiously, subsequently promoting the lactylation of histones. While previous studies have explored the impact of histone lactylation in modulating gene expression, the precise role of this epigenetic modification in regulating oncogenes is largely unchartered. In this study, using breast cancer cell lines and their mutants exhibiting lactate-deficient metabolome, we have identified that an enhanced rate of aerobic glycolysis supports c-Myc expression via promoter-level histone lactylation. Interestingly, c-Myc further transcriptionally upregulates serine/arginine splicing factor 10 ( SRSF10) to drive alternative splicing of MDM4 and Bcl-x in breast cancer cells. Moreover, our results reveal that restricting the activity of critical glycolytic enzymes affects the c-Myc-SRSF10 axis to subside the proliferation of breast cancer cells. Our findings provide novel insights into the mechanisms by which aerobic glycolysis influences alternative splicing processes that collectively contribute to breast tumorigenesis. Furthermore, we also envisage that chemotherapeutic interventions attenuating glycolytic rate can restrict breast cancer progression by impeding the c-Myc-SRSF10 axis.
Graphical abstract
Related collections
Most cited references70
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries
- Record: found
- Abstract: found
- Article: not found