- Record: found
- Abstract: found
- Article: found
Alterations of DNA damage response pathway: Biomarker and therapeutic strategy for cancer immunotherapy
review-article
Minlin Jiang
a
,
b ,
Keyi Jia
a
,
b ,
Lei Wang
a ,
Wei Li
a ,
Bin Chen
a ,
Yu Liu
a
,
b ,
Hao Wang
a
,
b ,
Sha Zhao
a ,
Yayi He
a
,
∗ ,
Caicun Zhou
a
,
∗
06 January 2021
DNA damage response,
DNA repair,
Immunotherapy,
Genomic instability,
Tumor microenvironment,
PD-1,
PD-L1,
cGAS–STING,
ATM, ataxia-telangiectasia mutated,
ATR, ataxia telangiectasia and Rad3 related,
BAP1, BRCA1-associated protein 1,
BER, base excision repair,
BRAF, v-RAF murine sarcoma viral oncogene homologue B,
BRCA, breast cancer susceptibility gene,
cGAS, cyclic GMP–AMP synthase,
CHEK, cell-cycle checkpoint kinase,
CHK1, checkpoint kinase 1,
DAMP, damage-associated molecular patterns,
DDR, DNA damage response,
DR, direct repair,
DSBs, double-strand breaks,
GSK3β, glycogen synthase kinase 3β,
HMGB1, high mobility group box-1,
HRR, homologous recombination repair,
ICI, immune checkpoint inhibitor,
IFNγ, interferon gamma,
IHC, immunohistochemistry,
IRF1, interferon regulatory factor 1,
JAK, Janus kinase,
MAD1, mitotic arrest deficient-like 1,
MGMT, O6-methylguanine methyltransferase,
MLH1, MutL homolog 1,
MMR, mismatch repair,
MNT, MAX network transcriptional repressor,
MSH2/6, MutS protein homologue-2/6,
MSI, microsatellite instability,
MUTYH, MutY homolog,
MyD88, myeloid differentiation factor 88,
NEK1, NIMA-related kinase 1,
NER, nucleotide excision repair,
NGS, next generation sequencing,
NHEJ, nonhomologous end-joining,
NIMA, never-in-mitosis A,
NSCLC, non-small cell lung cancer,
ORR, objective response rate,
OS, overall survival,
PALB2, partner and localizer of BRCA2,
PARP, poly-ADP ribose polymerase,
PCR, polymerase chain reaction,
PD-1, programmed death 1,
PD-L1, programmed death ligand 1,
PFS, progression-free survival,
RAD51C, RAD51 homolog C,
RB1, retinoblastoma 1,
RPA, replication protein A,
RSR, replication stress response,
SCNAs, somatic copy number alterations,
ssDNA, single-stranded DNA,
STAT, signal transducer and activator of transcription,
STING, stimulator of interferon genes,
TBK1, TANK-binding kinase 1,
TILs, tumor-infiltrating lymphocytes,
TLR4, Toll-like receptor 4,
TMB, tumor mutational burden,
TME, tumor microenvironment,
TP53, tumor protein P53,
TRIF, Toll-interleukin 1 receptor domain-containing adaptor inducing INF-β,
FDA, United State Food and Drug Administration,
XRCC4, X-ray repair cross complementing protein 4
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
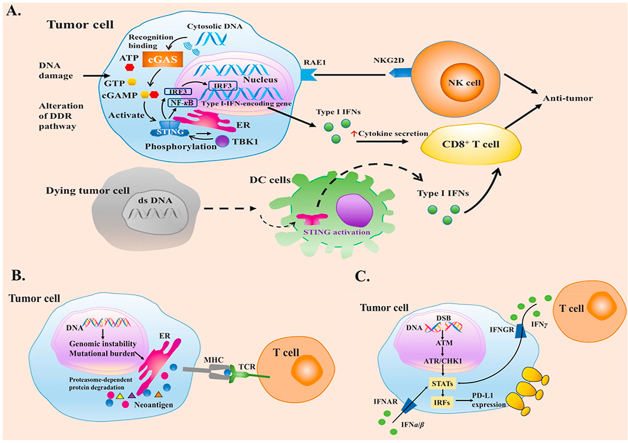
Genomic instability remains an enabling feature of cancer and promotes malignant transformation. Alterations of DNA damage response (DDR) pathways allow genomic instability, generate neoantigens, upregulate the expression of programmed death ligand 1 (PD-L1) and interact with signaling such as cyclic GMP–AMP synthase-stimulator of interferon genes (cGAS–STING) signaling. Here, we review the basic knowledge of DDR pathways, mechanisms of genomic instability induced by DDR alterations, impacts of DDR alterations on immune system, and the potential applications of DDR alterations as biomarkers and therapeutic targets in cancer immunotherapy.
Graphical abstract
Related collections
Most cited references106
- Record: found
- Abstract: found
- Article: not found
Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries
Ahmedin Jemal, Lindsey A Torre, Rebecca Siegel … (2018)
- Record: found
- Abstract: found
- Article: found
Hallmarks of Cancer: The Next Generation
Douglas Hanahan, Robert A. Weinberg (2011)
- Record: found
- Abstract: found
- Article: not found
Cancer statistics, 2020
Rebecca Siegel, Kimberly D Miller, Ahmedin Jemal (2020)
Author and article information
dna damage response,dna repair,immunotherapy,genomic instability,tumor microenvironment,pd-1,pd-l1,cgas–sting,atm, ataxia-telangiectasia mutated,atr, ataxia telangiectasia and rad3 related,bap1, brca1-associated protein 1,ber, base excision repair,braf, v-raf murine sarcoma viral oncogene homologue b,brca, breast cancer susceptibility gene,cgas, cyclic gmp–amp synthase,chek, cell-cycle checkpoint kinase,chk1, checkpoint kinase 1,damp, damage-associated molecular patterns,ddr, dna damage response,dr, direct repair,dsbs, double-strand breaks,gsk3β, glycogen synthase kinase 3β,hmgb1, high mobility group box-1,hrr, homologous recombination repair,ici, immune checkpoint inhibitor,ifnγ, interferon gamma,ihc, immunohistochemistry,irf1, interferon regulatory factor 1,jak, janus kinase,mad1, mitotic arrest deficient-like 1,mgmt, o6-methylguanine methyltransferase,mlh1, mutl homolog 1,mmr, mismatch repair,mnt, max network transcriptional repressor,msh2/6, muts protein homologue-2/6,msi, microsatellite instability,mutyh, muty homolog,myd88, myeloid differentiation factor 88,nek1, nima-related kinase 1,ner, nucleotide excision repair,ngs, next generation sequencing,nhej, nonhomologous end-joining,nima, never-in-mitosis a,nsclc, non-small cell lung cancer,orr, objective response rate,os, overall survival,palb2, partner and localizer of brca2,parp, poly-adp ribose polymerase,pcr, polymerase chain reaction,pd-1, programmed death 1,pd-l1, programmed death ligand 1,pfs, progression-free survival,rad51c, rad51 homolog c,rb1, retinoblastoma 1,rpa, replication protein a,rsr, replication stress response,scnas, somatic copy number alterations,ssdna, single-stranded dna,stat, signal transducer and activator of transcription,sting, stimulator of interferon genes,tbk1, tank-binding kinase 1,tils, tumor-infiltrating lymphocytes,tlr4, toll-like receptor 4,tmb, tumor mutational burden,tme, tumor microenvironment,tp53, tumor protein p53,trif, toll-interleukin 1 receptor domain-containing adaptor inducing inf-β,fda, united state food and drug administration,xrcc4, x-ray repair cross complementing protein 4
dna damage response, dna repair, immunotherapy, genomic instability, tumor microenvironment, pd-1, pd-l1, cgas–sting, atm, ataxia-telangiectasia mutated, atr, ataxia telangiectasia and rad3 related, bap1, brca1-associated protein 1, ber, base excision repair, braf, v-raf murine sarcoma viral oncogene homologue b, brca, breast cancer susceptibility gene, cgas, cyclic gmp–amp synthase, chek, cell-cycle checkpoint kinase, chk1, checkpoint kinase 1, damp, damage-associated molecular patterns, ddr, dna damage response, dr, direct repair, dsbs, double-strand breaks, gsk3β, glycogen synthase kinase 3β, hmgb1, high mobility group box-1, hrr, homologous recombination repair, ici, immune checkpoint inhibitor, ifnγ, interferon gamma, ihc, immunohistochemistry, irf1, interferon regulatory factor 1, jak, janus kinase, mad1, mitotic arrest deficient-like 1, mgmt, o6-methylguanine methyltransferase, mlh1, mutl homolog 1, mmr, mismatch repair, mnt, max network transcriptional repressor, msh2/6, muts protein homologue-2/6, msi, microsatellite instability, mutyh, muty homolog, myd88, myeloid differentiation factor 88, nek1, nima-related kinase 1, ner, nucleotide excision repair, ngs, next generation sequencing, nhej, nonhomologous end-joining, nima, never-in-mitosis a, nsclc, non-small cell lung cancer, orr, objective response rate, os, overall survival, palb2, partner and localizer of brca2, parp, poly-adp ribose polymerase, pcr, polymerase chain reaction, pd-1, programmed death 1, pd-l1, programmed death ligand 1, pfs, progression-free survival, rad51c, rad51 homolog c, rb1, retinoblastoma 1, rpa, replication protein a, rsr, replication stress response, scnas, somatic copy number alterations, ssdna, single-stranded dna, stat, signal transducer and activator of transcription, sting, stimulator of interferon genes, tbk1, tank-binding kinase 1, tils, tumor-infiltrating lymphocytes, tlr4, toll-like receptor 4, tmb, tumor mutational burden, tme, tumor microenvironment, tp53, tumor protein p53, trif, toll-interleukin 1 receptor domain-containing adaptor inducing inf-β, fda, united state food and drug administration, xrcc4, x-ray repair cross complementing protein 4