- Record: found
- Abstract: found
- Article: found
Genomic Characterization of Lactobacillus delbrueckii TUA4408L and Evaluation of the Antiviral Activities of its Extracellular Polysaccharides in Porcine Intestinal Epithelial Cells
Read this article at
Abstract
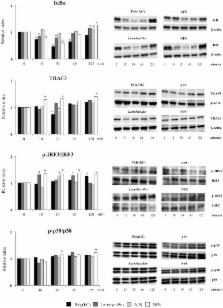
In lactic acid bacteria, the synthesis of exopolysaccharides (EPS) has been associated with some favorable technological properties as well as health-promoting benefits. Research works have shown the potential of EPS produced by lactobacilli to differentially modulate immune responses. However, most studies were performed in immune cells and few works have concentrated in the immunomodulatory activities of EPS in non-immune cells such as intestinal epithelial cells. In addition, the cellular and molecular mechanisms involved in the immunoregulatory effects of EPS have not been studied in detail. In this work, we have performed a genomic characterization of Lactobacillus delbrueckii subsp. delbrueckii TUA4408L and evaluated the immunomodulatory and antiviral properties of its acidic (APS) and neutral (NPS) EPS in porcine intestinal epithelial (PIE) cells. Whole genome sequencing allowed the analysis of the general features of L. delbrueckii TUA4408L genome as well as the characterization of its EPS genes. A typical EPS gene cluster was found in the TUA4408L genome consisting in five highly conserved genes epsA- E, and a variable region, which includes the genes for the polymerase wzy, the flippase wzx, and seven glycosyltransferases. In addition, we demonstrated here for the first time that L. delbrueckii TUA4408L and its EPS are able to improve the resistance of PIE cells against rotavirus infection by reducing viral replication and regulating inflammatory response. Moreover, studies in PIE cells demonstrated that the TUA4408L strain and its EPS differentially modulate the antiviral innate immune response triggered by the activation of Toll-like receptor 3 (TLR3). L. delbrueckii TUA4408L and its EPS are capable of increasing the activation of interferon regulatory factor (IRF)-3 and nuclear factor κB (NF-κB) signaling pathways leading to an improved expression of the antiviral factors interferon (IFN)-β, Myxovirus resistance gene A (MxA) and RNaseL.
Related collections
Most cited references31

- Record: found
- Abstract: found
- Article: found
IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets


- Record: found
- Abstract: found
- Article: found