- Record: found
- Abstract: found
- Article: found
Characterization of a NDM-1- Encoding Plasmid pHFK418-NDM From a Clinical Proteus mirabilis Isolate Harboring Two Novel Transposons, Tn 6624 and Tn 6625
Read this article at
Abstract
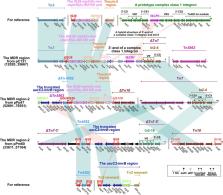
Acquisition of the bla NDM– 1 gene by Proteus mirabilis is a concern because it already has intrinsic resistance to polymyxin E and tigecycline antibiotics. Here, we describe a P. mirabilis isolate that carries a pPrY2001-like plasmid (pHFK418-NDM) containing a bla NDM– 1 gene. The pPrY2001-like plasmid, pHFK418-NDM, was first reported in China. The pHFK418-NDM plasmid was sequenced using a hybrid approach based on Illumina and MinION platforms. The sequence of pHFK418-NDM was compared with those of the six other pPrY2001-like plasmids deposited in GenBank. We found that the multidrug-resistance encoding region of pHFK418-NDM contains ΔTn 10 and a novel transposon Tn 6625. Tn 6625 consists of ΔTn 1696, Tn 6260, In251, ΔTn 125 (carrying bla NDM– 1), ΔTn 2670, and a novel mph(E)-harboring transposon Tn 6624. In251 was first identified in a clinical isolate, suggesting that it has been transferred efficiently from environmental organisms to clinical isolates. Genomic comparisons of all these pPrY2001-like plasmids showed that their relatively conserved backbones could integrate the numerous and various accessory modules carrying multifarious antibiotic resistance genes. Our results provide a greater depth of insight into the horizontal transfer of resistance genes and add interpretive value to the genomic diversity and evolution of pPrY2001-like plasmids.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
ISfinder: the reference centre for bacterial insertion sequences
- Record: found
- Abstract: found
- Article: not found
Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae.
- Record: found
- Abstract: found
- Article: not found