- Record: found
- Abstract: found
- Article: found
Complete Genomes of Symbiotic Cyanobacteria Clarify the Evolution of Vanadium-Nitrogenase
Read this article at
Abstract
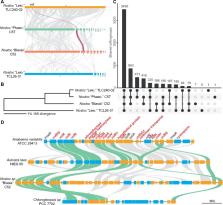
Plant endosymbiosis with nitrogen-fixing cyanobacteria has independently evolved in diverse plant lineages, offering a unique window to study the evolution and genetics of plant–microbe interaction. However, very few complete genomes exist for plant cyanobionts, and therefore little is known about their genomic and functional diversity. Here, we present four complete genomes of cyanobacteria isolated from bryophytes. Nanopore long-read sequencing allowed us to obtain circular contigs for all the main chromosomes and most of the plasmids. We found that despite having a low 16S rRNA sequence divergence, the four isolates exhibit considerable genome reorganizations and variation in gene content. Furthermore, three of the four isolates possess genes encoding vanadium (V)-nitrogenase ( vnf), which is uncommon among diazotrophs and has not been previously reported in plant cyanobionts. In two cases, the vnf genes were found on plasmids, implying possible plasmid-mediated horizontal gene transfers. Comparative genomic analysis of vnf-containing cyanobacteria further identified a conserved gene cluster. Many genes in this cluster have not been functionally characterized and would be promising candidates for future studies to elucidate V-nitrogenase function and regulation.
Related collections
Most cited references23
- Record: found
- Abstract: found
- Article: not found
Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps.

- Record: found
- Abstract: found
- Article: found
Fern genomes elucidate land plant evolution and cyanobacterial symbioses

- Record: found
- Abstract: found
- Article: found