- Record: found
- Abstract: found
- Article: not found
Fat cells reactivate quiescent neuroblasts via TOR and glial Insulin relays in Drosophila
Abstract
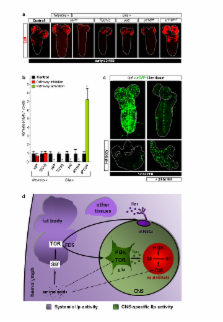
Many stem, progenitor and cancer cells undergo periods of mitotic quiescence from which they can be reactivated 1- 5 . The signals triggering entry into and exit from this reversible dormant state are not well understood. In the developing Drosophila central nervous system (CNS), multipotent self-renewing progenitors called neuroblasts 6- 9 undergo quiescence in a stereotypical spatiotemporal pattern 10 . Entry into quiescence is regulated by Hox proteins and an internal neuroblast timer 11- 13 . Exit from quiescence (reactivation) is subject to a nutritional checkpoint requiring dietary amino acids 14 . Organ co-cultures also implicate an unidentified signal from an adipose/hepatic-like tissue called fat body 14 . Here, we provide in vivo evidence that Slimfast amino-acid sensing and Target-of-Rapamycin (TOR) signalling 15 activate a fat-body derived signal (FDS) required for neuroblast reactivation. Downstream of the FDS, Insulin-like receptor (InR) signalling and the Phosphatidylinositol 3-Kinase (PI3K)/TOR network are required in neuroblasts for exit from quiescence. We demonstrate that nutritionally regulated glial cells provide the source of Insulin-like Peptides (Ilps) relevant for timely neuroblast reactivation but not for overall larval growth. Conversely, Ilps secreted into the hemolymph by median neurosecretory cells (mNSCs) systemically control organismal size 16- 18 but do not reactivate neuroblasts. Drosophila thus contains two segregated Ilp pools, one regulating proliferation within the CNS and the other controlling tissue growth systemically. Together, our findings support a model in which amino acids trigger the cell cycle re-entry of neural progenitors via a fat body→glia→neuroblasts relay. This mechanism highlights that dietary nutrients and remote organs, as well as local niches, are key regulators of transitions in stem-cell behaviour.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Structure and function of the feed-forward loop network motif.
- Record: found
- Abstract: found
- Article: not found
Slit is the midline repellent for the robo receptor in Drosophila.
- Record: found
- Abstract: found
- Article: not found
Molecular mechanisms of metabolic regulation by insulin in Drosophila.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.