- Record: found
- Abstract: found
- Article: found
How Attractive Is the Girl Next Door? An Assessment of Spatial Mate Acquisition and Paternity in the Solitary Cape Dune Mole-Rat, Bathyergus suillus
Read this article at
Abstract
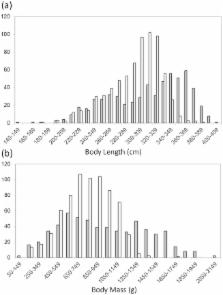
Behavioural observations of reproduction and mate choice in wild fossorial rodents are extremely limited and consequently indirect methods are typically used to infer mating strategies. We use a combination of morphological, reproductive, spatial, and genetic data to investigate the reproductive strategy of a solitary endemic species, the Cape dune mole-rat Bathyergus suillus. These data provide the first account on the population dynamics of this species. Marked sexual dimorphism was apparent with males being both significantly larger and heavier than females. Of all females sampled 36% had previously reproduced and 12% were pregnant at the time of capture. Post-partum sex ratio was found to be significantly skewed in favour of females. The paternity of fifteen litters (n = 37) was calculated, with sires assigned to progeny using both categorical and full probability methods, and including a distance function. The maximum distance between progeny and a putative sire was determined as 2149 m with males moving between sub-populations. We suggest that above-ground movement should not be ignored in the consideration of mate acquisition behaviour of subterranean mammals. Estimated levels of multiple paternity were shown to be potentially as high as 26%, as determined using sibship and sire assignment methods. Such high levels of multiple paternity have not been found in other solitary mole-rat species. The data therefore suggest polyandry with no evidence as yet for polygyny.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: not found
Statistical confidence for likelihood-based paternity inference in natural populations.
- Record: found
- Abstract: found
- Article: not found
Sibship reconstruction from genetic data with typing errors.
- Record: found
- Abstract: not found
- Article: not found