- Record: found
- Abstract: found
- Article: found
Itaconate-producing neutrophils regulate local and systemic inflammation following trauma

Read this article at
Abstract
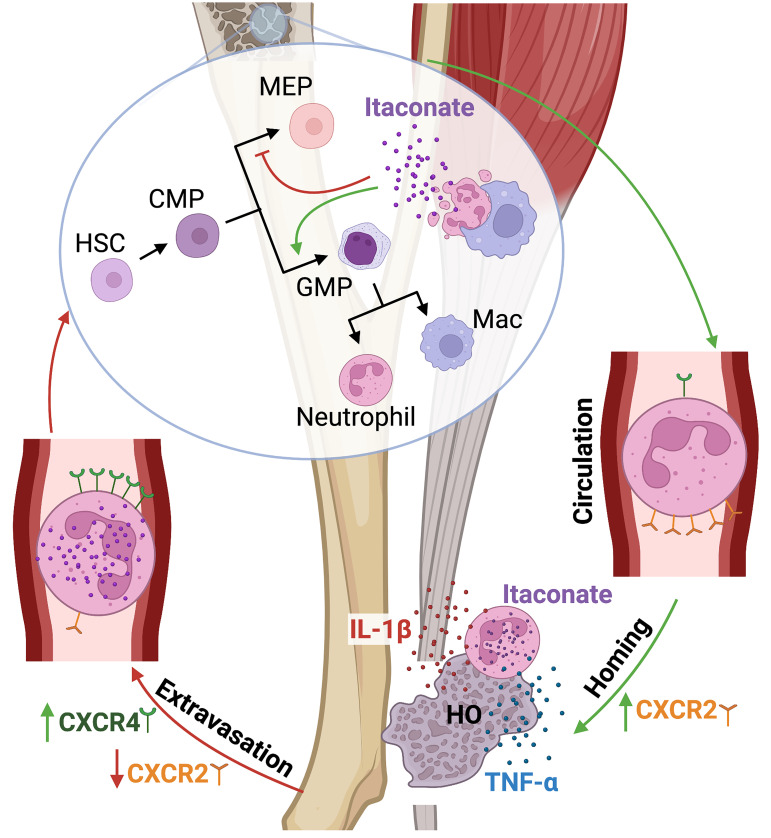
Modulation of the immune response to initiate and halt the inflammatory process occurs both at the site of injury as well as systemically. Due to the evolving role of cellular metabolism in regulating cell fate and function, tendon injuries that undergo normal and aberrant repair were evaluated by metabolic profiling to determine its impact on healing outcomes. Metabolomics revealed an increasing abundance of the immunomodulatory metabolite itaconate within the injury site. Subsequent single-cell RNA-Seq and molecular and metabolomic validation identified a highly mature neutrophil subtype, not macrophages, as the primary producers of itaconate following trauma. These mature itaconate-producing neutrophils were highly inflammatory, producing cytokines that promote local injury fibrosis before cycling back to the bone marrow. In the bone marrow, itaconate was shown to alter hematopoiesis, skewing progenitor cells down myeloid lineages, thereby regulating systemic inflammation. Therapeutically, exogenous itaconate was found to reduce injury-site inflammation, promoting tenogenic differentiation and impairing aberrant vascularization with disease-ameliorating effects. These results present an intriguing role for cycling neutrophils as a sensor of inflammation induced by injury — potentially regulating immune cell production in the bone marrow through delivery of endogenously produced itaconate — and demonstrate a therapeutic potential for exogenous itaconate following tendon injury
Abstract
Related collections
Most cited references62

- Record: found
- Abstract: found
- Article: found
Integrated analysis of multimodal single-cell data

- Record: found
- Abstract: found
- Article: found
Inference and analysis of cell-cell communication using CellChat
- Record: found
- Abstract: found
- Article: not found