- Record: found
- Abstract: found
- Article: found
Genome-wide identification and expression profile of GhGRF gene family in Gossypium hirsutum L.
Read this article at
Abstract
Background
Cotton is the primary source of renewable natural fiber in the textile industry and an important biodiesel crop. Growth regulating factors ( GRFs) are involved in regulating plant growth and development.
Results
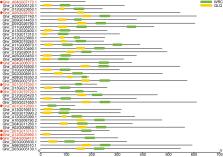
Chromosomal location information revealed an uneven distribution of GhGRF genes, with maximum genes on chromosomes A02, A05, and A12 from the At sub-genome and their corresponding D05 and D12 from the Dt sub-genome. In the phylogenetic tree, 35 GRF genes were divided into five groups, including G1, G2, G3, G4, and G5. The majority of GhGRF genes have two to three introns and three to four exons, and their deduced proteins contained conserved QLQ and WRC domains in the N-terminal end of GRFs in Arabidopsis and rice. Sequence logos revealed that GRF genes were highly conserved during the long-term evolutionary process. The CDS of the GhGRF gene can complement MiRNA396a. Moreover, most GhGRF genes transcripts developed high levels of ovules and fibers. Analyses of promoter cis-elements and expression patterns indicated that GhGRF genes play an essential role in regulating plant growth and development by coordinating the internal and external environment and multiple hormone signaling pathways. Our analysis indicated that GhGRFs are ideal target genes with significant potential for improving the molecular structure of cotton.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
WebLogo: a sequence logo generator.
- Record: found
- Abstract: found
- Article: not found
MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.
- Record: found
- Abstract: found
- Article: not found