- Record: found
- Abstract: not found
- Article: not found
Performance of Rapid Antigen Tests to Detect Symptomatic and Asymptomatic SARS-CoV-2 Infection : A Prospective Cohort Study
Apurv Soni
1 ,
Carly Herbert
2 ,
Honghuang Lin
3 ,
Yi Yan
4 ,
Caitlin Pretz
2 ,
Pamela Stamegna
2 ,
Biqi Wang
3 ,
Taylor Orwig
2 ,
Colton Wright
2 ,
Seanan Tarrant
2 ,
Stephanie Behar
2 ,
Thejas Suvarna
5 ,
Summer Schrader
5 ,
Emma Harman
5 ,
Chris Nowak
5 ,
Vik Kheterpal
5 ,
Lokinendi V. Rao
6 ,
Lisa Cashman
6 ,
Elizabeth Orvek
7 ,
Didem Ayturk
7 ,
Laura Gibson
8 ,
Adrian Zai
7 ,
Steven Wong
7 ,
Peter Lazar
7 ,
Ziyue Wang
9 ,
Andreas Filippaios
2 ,
Bruce Barton
7 ,
Chad J. Achenbach
10 ,
Robert L. Murphy
10 ,
Matthew L. Robinson
11 ,
Yukari C. Manabe
11 ,
Shishir Pandey
2 ,
Andres Colubri
12 ,
Laurel O’Connor
13 ,
Stephenie C. Lemon
7 ,
Nisha Fahey
14 ,
Katherine L. Luzuriaga
15 ,
Nathaniel Hafer
15 ,
Kristian Roth
4 ,
Toby Lowe
4 ,
Timothy Stenzel
16 ,
William Heetderks
17 ,
John Broach
13 ,
David D. McManus
18
July 04 2023
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
<p xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="first" id="d8299052e770">This
prospective study evaluated the performance of rapid antigen tests for detection
of SARS-CoV-2 among symptomatic and asymptomatic participants. Participants who were
asymptomatic and negative for SARS-CoV-2 on study day 1 completed rapid antigen tests
and RT-PCR testing for SARS-CoV-2 every 48 hours for 15 days.
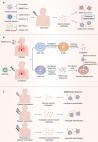
</p><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="fig panel" id="fva1-M230385">
<a class="named-anchor" id="fva1-M230385">
<!--
named anchor
-->
</a>
<div class="figure-container so-text-align-c">
<img alt="" class="figure" src="/document_file/96ade31c-ed83-4189-8ec2-4a6ded166e27/PubMedCentral/image/aim-olf-M230385-AIME202307180-M230385_visual-abstract"/>
</div>
<div class="panel-content">
<div class="label">Visual Abstract.</div>
<div class="caption" id="d8299052e776">
<strong>
<span class="fig-title">Performance of Rapid Antigen Tests to Detect Symptomatic and
Asymptomatic SARS-CoV-2
Infection
</span>
</strong>
<p id="d8299052e779">This prospective study evaluated the performance of rapid antigen
tests for detection
of SARS-CoV-2 among symptomatic and asymptomatic participants. Participants who were
asymptomatic and negative for SARS-CoV-2 on study day 1 completed rapid antigen tests
and RT-PCR testing for SARS-CoV-2 every 48 hours for 15 days.
</p>
</div>
</div>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e783">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e784">Background:</h5>
<p id="d8299052e786">The performance of rapid antigen tests (Ag-RDTs) for screening
asymptomatic and symptomatic
persons for SARS-CoV-2 is not well established.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e788">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e789">Objective:</h5>
<p id="d8299052e791">To evaluate the performance of Ag-RDTs for detection of SARS-CoV-2
among symptomatic
and asymptomatic participants.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e793">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e794">Design:</h5>
<p id="d8299052e796">This prospective cohort study enrolled participants between October
2021 and January
2022. Participants completed Ag-RDTs and reverse transcriptase polymerase chain reaction
(RT-PCR) testing for SARS-CoV-2 every 48 hours for 15 days.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e798">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e799">Setting:</h5>
<p id="d8299052e801">Participants were enrolled digitally throughout the mainland
United States. They self-collected
anterior nasal swabs for Ag-RDTs and RT-PCR testing. Nasal swabs for RT-PCR were shipped
to a central laboratory, whereas Ag-RDTs were done at home.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e803">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e804">Participants:</h5>
<p id="d8299052e806">Of 7361 participants in the study, 5353 who were asymptomatic
and negative for SARS-CoV-2
on study day 1 were eligible. In total, 154 participants had at least 1 positive RT-PCR
result.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e808">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e809">Measurements:</h5>
<p id="d8299052e811">The sensitivity of Ag-RDTs was measured on the basis of testing
once (same-day), twice
(after 48 hours), and thrice (after a total of 96 hours). The analysis was repeated
for different days past index PCR positivity (DPIPPs) to approximate real-world scenarios
where testing initiation may not always coincide with DPIPP 0. Results were stratified
by symptom status.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e813">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e814">Results:</h5>
<p id="d8299052e816">Among 154 participants who tested positive for SARS-CoV-2, 97
were asymptomatic and
57 had symptoms at infection onset. Serial testing with Ag-RDTs twice 48 hours apart
resulted in an aggregated sensitivity of 93.4% (95% CI, 90.4% to 95.9%) among symptomatic
participants on DPIPPs 0 to 6. When singleton positive results were excluded, the
aggregated sensitivity on DPIPPs 0 to 6 for 2-time serial testing among asymptomatic
participants was lower at 62.7% (CI, 57.0% to 70.5%), but it improved to 79.0% (CI,
70.1% to 87.4%) with testing 3 times at 48-hour intervals.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e818">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e819">Limitation:</h5>
<p id="d8299052e821">Participants tested every 48 hours; therefore, these data cannot
support conclusions
about serial testing intervals shorter than 48 hours.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e823">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e824">Conclusion:</h5>
<p id="d8299052e826">The performance of Ag-RDTs was optimized when asymptomatic participants
tested 3 times
at 48-hour intervals and when symptomatic participants tested 2 times separated by
48 hours.
</p>
</div><div xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" class="section">
<a class="named-anchor" id="d8299052e828">
<!--
named anchor
-->
</a>
<h5 class="section-title" id="d8299052e829">Primary Funding Source:</h5>
<p id="d8299052e831">National Institutes of Health RADx Tech program.</p>
</div>
Related collections
Most cited references17
- Record: found
- Abstract: found
- Article: not found
Evaluation of rapid antigen test for detection of SARS-CoV-2 virus
Gannon Mak, Peter Cheng, Stephen Lau … (2020)
- Record: found
- Abstract: found
- Article: found
SARS-CoV-2 Omicron variant: recent progress and future perspectives
- Record: found
- Abstract: found
- Article: not found
Scaling up COVID-19 rapid antigen tests: promises and challenges
Rosanna Peeling, Piero L. Olliaro, Debrah I Boeras … (2021)
