- Record: found
- Abstract: found
- Article: found
Tnfrsf4-expressing regulatory T cells promote immune escape of chronic myeloid leukemia stem cells

Read this article at
Abstract
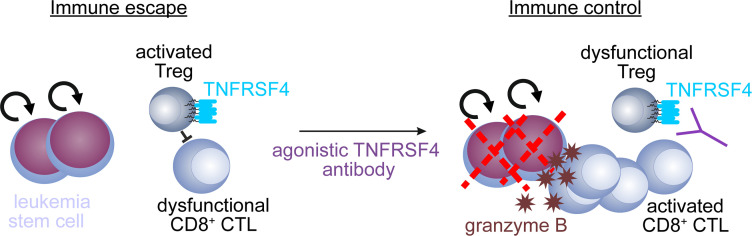
Leukemia stem cells (LSCs) promote the disease and seem resistant to therapy and immune control. Why LSCs are selectively resistant against elimination by CD8 + cytotoxic T cells (CTLs) is still unknown. In this study, we demonstrate that LSCs in chronic myeloid leukemia (CML) can be recognized and killed by CD8 + CTLs in vitro. However, Tregs, which preferentially localized close to CD8 + CTLs in CML BM, protected LSCs from MHC class I–dependent CD8 + CTL–mediated elimination in vivo. BM Tregs in CML were characterized by the selective expression of tumor necrosis factor receptor 4 (Tnfrsf4). Stimulation of Tnfrsf4 signaling did not deplete Tregs but reduced the capacity of Tregs to protect LSCs from CD8 + CTL–mediated killing. In the BM of newly diagnosed CML patients, TNFRSF4 mRNA levels were significantly increased and correlated with the expression of the Treg-restricted transcription factor FOXP3. Overall, these results identify Tregs as key regulators of immune escape of LSCs and TNFRSF4 as a potential target to reduce the function of Tregs and boost antileukemic immunity in CML.
Abstract
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support.
- Record: found
- Abstract: found
- Article: not found
Signatures of mutational processes in human cancer

- Record: found
- Abstract: found
- Article: found