- Record: found
- Abstract: found
- Article: found
Sycp2 is essential for synaptonemal complex assembly, early meiotic recombination and homologous pairing in zebrafish spermatocytes
Read this article at
Abstract
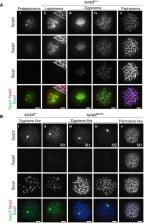
Meiotic recombination is essential for faithful segregation of homologous chromosomes during gametogenesis. The progression of recombination is associated with dynamic changes in meiotic chromatin structures. However, whether Sycp2, a key structural component of meiotic chromatin, is required for the initiation of meiotic recombination is still unclear in vertebrates. Here, we describe that Sycp2 is required for assembly of the synaptonemal complex and early meiotic events in zebrafish spermatocytes. Our genetic screening by N-ethyl-N-nitrosourea mutagenesis revealed that ietsugu ( its), a mutant zebrafish line with an aberrant splice site in the sycp2 gene, showed a defect during meiotic prophase I. The its mutation appeared to be a hypomorphic mutation compared to sycp2 knockout mutations generated by TALEN mutagenesis. Taking advantage of these sycp2 hypomorphic and knockout mutant lines, we demonstrated that Sycp2 is required for the assembly of the synaptonemal complex that is initiated in the vicinity of telomeres in wild-type zebrafish spermatocytes. Accordingly, homologous pairing, the foci of the meiotic recombinases Dmc1/Rad51 and RPA, and γH2AX signals were largely diminished in sycp2 knockout spermatocytes. Taken together, our data indicate that Sycp2 plays a critical role in not only the assembly of the synaptonemal complex, but also early meiotic recombination and homologous pairing, in vertebrates.
Author summary
Meiosis is an essential type of cell division whose purpose is to produce gametes, such as eggs and sperm, for sexually reproducing eukaryotes. Most cells in the human body are diploid cells containing two homologous copies of each chromosome. Meiosis halves the genetic contents of diploid cells and produces haploid gametes with a single copy of each homologous chromosome pair. The faithful transmission of chromosomes is one of the main tasks in meiosis, since extra or missing chromosomes are common causes of genetic disorders, birth defects and infertility. Prior to proper segregation, homologs must pair up and be physically connected. This is achieved by homologous recombination initiated by programmed DNA double-strand breaks. Since it can be deleterious to cells, this initiation step is highly regulated by factors that are not fully understood. In this study, we found that Sycp2, a structural component of meiotic chromatin in metazoans, is essential for the initiation of homologous recombination, meiotic chromatin organization, and homologous pairing in zebrafish. These findings support recent biochemical studies that Sycp2 and Sycp3 serve as a vertebrate counterpart of S. cerevisiae Red1.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Efficient In Vivo Genome Editing Using RNA-Guided Nucleases

- Record: found
- Abstract: found
- Article: found
Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting
- Record: found
- Abstract: found
- Article: not found