- Record: found
- Abstract: found
- Article: found
The interferon-β/STAT1 axis drives the collective invasion of skin squamous cell carcinoma with sealed intercellular spaces
Read this article at
Abstract
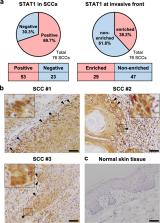
The process by which cancer cells invade as a cell cluster, known as collective invasion, is associated with metastasis and worse prognosis of cancer patients; therefore, inhibition of collective invasion is considered to improve cancer treatment. However, the cellular characteristics responsible for collective invasion remain largely unknown. Here, we successfully established subclones with various invasive potentials derived from human skin squamous carcinoma cells. The cell cluster of the highly invasive subclone had a hermetically sealed and narrow intercellular space. Interferon-β was localized to the sealed intercellular spaces, leading to collective invasion via the activation of signal transducer and activator of transcription 1 (STAT1). On the other hand, interferon-β was not localized to non-sealed and wide intercellular spaces of the cell cluster of low-invasive subclone with deficient STAT1 activity. In the mixed cell cluster of high- and low-invasive subclones, the high-invasive sub-clonal cells were located at the invasive front of the invasive protrusion, leading to collective invasion by the low-invasive sub-clonal cells. Tissue microarray analysis of human skin squamous cell carcinoma (SCC) also showed enrichment of STAT1 in the invasive front of SCCs. These findings indicate that the intercellular structure controls the potential for collective invasion via STAT1 regulation in SCC.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Regulation of type I interferon responses.
- Record: found
- Abstract: found
- Article: not found
Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis.
- Record: found
- Abstract: found
- Article: not found