- Record: found
- Abstract: found
- Article: found
The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity
Read this article at
Abstract
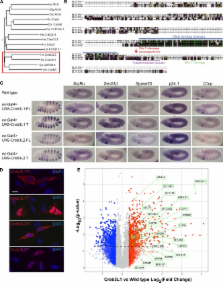
CrebA up-regulates expression of both the general protein machinery required in all cells for secretion and genes encoding cell type–specific secreted components.
Abstract
Secretion occurs in all cells, with relatively low levels in most cells and extremely high levels in specialized secretory cells, such as those of the pancreas, salivary, and mammary glands. How secretory capacity is selectively up-regulated in specialized secretory cells is unknown. Here, we find that the CrebA/Creb3-like family of bZip transcription factors functions to up-regulate expression of both the general protein machinery required in all cells for secretion and of cell type–specific secreted proteins. Drosophila CrebA directly binds the enhancers of secretory pathway genes and is both necessary and sufficient to activate expression of every secretory pathway component gene examined thus far. Microarray profiling reveals that CrebA also up-regulates expression of genes encoding cell type–specific secreted components. Finally, we found that the human CrebA orthologues, Creb3L1 and Creb3L2, have the ability to up-regulate the secretory pathway in nonsecretory cell types.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
DAVID: Database for Annotation, Visualization, and Integrated Discovery.
- Record: found
- Abstract: found
- Article: not found
Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response.
- Record: found
- Abstract: found
- Article: not found