- Record: found
- Abstract: found
- Article: found
Identification of key interferon-stimulated genes for indicating the condition of patients with systemic lupus erythematosus
Read this article at
Abstract
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease with highly heterogeneous clinical symptoms and severity. There is complex pathogenesis of SLE, one of which is IFNs overproduction and downstream IFN-stimulated genes (ISGs) upregulation. Identifying the key ISGs differentially expressed in peripheral blood mononuclear cells (PBMCs) of patients with SLE and healthy people could help to further understand the role of the IFN pathway in SLE and discover potential diagnostic biomarkers.
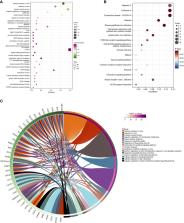
The differentially expressed ISGs (DEISG) in PBMCs of SLE patients and healthy persons were screened from two datasets of the Gene Expression Omnibus (GEO) database. A total of 67 DEISGs, including 6 long noncoding RNAs (lncRNAs) and 61 messenger RNAs (mRNAs) were identified by the “DESeq2” R package. According to Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, those DEISGs were mainly concentrated in the response to virus and immune system processes. Protein-protein interaction (PPI) network showed that most of these DEISGs could interact strongly with each other. Then, IFIT1, RSAD2, IFIT3, USP18, ISG15, OASL, MX1, OAS2, OAS3, and IFI44 were considered to be hub ISGs in SLE by “MCODE” and “Cytohubba” plugins of Cytoscape, Moreover, the results of expression correlation suggested that 3 lncRNAs (NRIR, FAM225A, and LY6E-DT) were closely related to the IFN pathway.
The lncRNA NRIR and mRNAs (RSAD2, USP18, IFI44, and ISG15) were selected as candidate ISGs for verification. RT-qPCR results showed that PBMCs from SLE patients had substantially higher expression levels of 5 ISGs compared to healthy controls (HCs). Additionally, statistical analyses revealed that the expression levels of these ISGs were strongly associated to various clinical symptoms, including thrombocytopenia and facial erythema, as well as laboratory indications, including the white blood cell (WBC) count and levels of autoantibodies. The Receiver Operating Characteristic (ROC) curve demonstrated that the IFI44, USP18, RSAD2, and IFN score had good diagnostic capabilities of SLE.
According to our study, SLE was associated with ISGs including NRIR, RSAD2, USP18, IFI44, and ISG15, which may contribute to the future diagnosis and new personalized targeted therapies.
Related collections
Most cited references63

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: not found
- Article: not found
Updating the American college of rheumatology revised criteria for the classification of systemic lupus erythematosus
- Record: found
- Abstract: found
- Article: not found