- Record: found
- Abstract: found
- Article: found
Q&A: Array tomography
Read this article at
Abstract
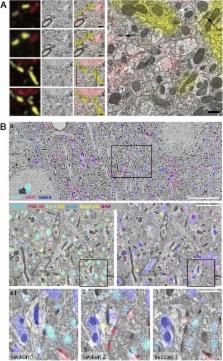
Array tomography encompasses light and electron microscopy modalities that offer unparalleled opportunities to explore three-dimensional cellular architectures in extremely fine structural and molecular detail. Fluorescence array tomography achieves much higher resolution and molecular multiplexing than most other fluorescence microscopy methods, while electron array tomography can capture three-dimensional ultrastructure much more easily and rapidly than traditional serial-section electron microscopy methods. A correlative fluorescence/electron microscopy mode of array tomography furthermore offers a unique capacity to merge the molecular discrimination strengths of multichannel fluorescence microscopy with the ultrastructural imaging strengths of electron microscopy. This essay samples the first decade of array tomography, highlighting applications in neuroscience.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors
- Record: found
- Abstract: found
- Article: not found
Protein localization in electron micrographs using fluorescence nanoscopy
- Record: found
- Abstract: found
- Article: not found