- Record: found
- Abstract: found
- Article: found
The melatonin receptor 1B gene links circadian rhythms and type 2 diabetes mellitus: an evolutionary story
Read this article at
Abstract
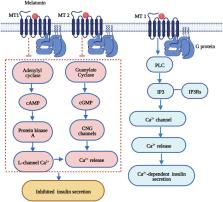
Disturbed circadian rhythms have been a risk factor for type 2 diabetes mellitus (T2DM). Melatonin is the major chronobiotic hormone regulating both circadian rhythm and glucose homeostasis. The rs10830963 (G allele) of the melatonin receptor 1B ( MTNR1B) gene has the strongest genetic associations with T2DM according to several genome-wide association studies. The MTNR1B rs10830963 G allele is also associated with disturbed circadian phenotypes and altered melatonin secretion, both factors that can elevate the risk of diabetes. Furthermore, evolutionary studies implied the presence of selection pressure and ethnic diversity in MTNR1B, which was consistent with the “thrifty gene” hypothesis in T2DM. The rs10830963 G risk allele is associated with delayed melatonin secretion onset in dim-light and prolonged duration of peak melatonin. This delayed melatonin secretion may help human ancestors adapt to famine or food shortages during long nights and early mornings and avoid nocturnal hypoglycemia but confers susceptibility to T2DM due to adequate energy intake in modern society. We provide new insight into the role of MTNR1B variants in T2DM via disturbed circadian rhythms from the perspective of the “thrifty gene” hypothesis; these data indicate a novel target for the prevention and treatment of susceptible populations with the thrifty genotype.
Related collections
Most cited references234
- Record: found
- Abstract: found
- Article: not found
Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis.
- Record: found
- Abstract: found
- Article: not found
Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps
- Record: found
- Abstract: found
- Article: not found