- Record: found
- Abstract: found
- Article: found
Plant Hormonomics: A Key Tool for Deep Physiological Phenotyping to Improve Crop Productivity
Read this article at
Abstract
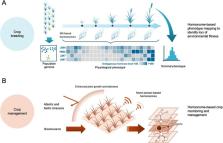
Agriculture is particularly vulnerable to climate change. To cope with the risks posed by climate-related stressors to agricultural production, global population growth, and changes in food preferences, it is imperative to develop new climate-smart crop varieties with increased yield and environmental resilience. Molecular genetics and genomic analyses have revealed that allelic variations in genes involved in phytohormone-mediated growth regulation have greatly improved productivity in major crops. Plant science has remarkably advanced our understanding of the molecular basis of various phytohormone-mediated events in plant life. These findings provide essential information for improving the productivity of crops growing in changing climates. In this review, we highlight the recent advances in plant hormonomics (multiple phytohormone profiling) and discuss its application to crop improvement. We present plant hormonomics as a key tool for deep physiological phenotyping, focusing on representative plant growth regulators associated with the improvement of crop productivity. Specifically, we review advanced methodologies in plant hormonomics, highlighting mass spectrometry- and nanosensor-based plant hormone profiling techniques. We also discuss the applications of plant hormonomics in crop improvement through breeding and agricultural management practices.
Related collections
Most cited references225
- Record: found
- Abstract: found
- Article: not found
mRNA-Seq whole-transcriptome analysis of a single cell.
- Record: found
- Abstract: found
- Article: not found