- Record: found
- Abstract: found
- Article: found
Phylogeographic Insights into a Peripheral Refugium: The Importance of Cumulative Effect of Glaciation on the Genetic Structure of Two Endemic Plants
Read this article at
Abstract
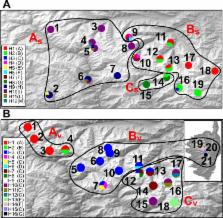
Quaternary glaciations and mostly last glacial maximum have shaped the contemporary distribution of many species in the Alps. However, in the Maritime and Ligurian Alps a more complex picture is suggested by the presence of many Tertiary paleoendemisms and by the divergence time between lineages in one endemic species predating the Late Pleistocene glaciation. The low number of endemic species studied limits the understanding of the processes that took place within this region. We used species distribution models and phylogeographical methods to infer glacial refugia and to reconstruct the phylogeographical pattern of Silene cordifolia All. and Viola argenteria Moraldo & Forneris. The predicted suitable area for last glacial maximum roughly fitted current known distribution. Our results suggest that separation of the major clades predates the last glacial maximum and the following repeated glacial and interglacial periods probably drove differentiations. The complex phylogeographical pattern observed in the study species suggests that both populations and genotypes extinction was minimal during the last glacial maximum, probably due to the low impact of glaciations and to topographic complexity in this area. This study underlines the importance of cumulative effect of previous glacial cycles in shaping the genetic structure of plant species in Maritime and Ligurian Alps, as expected for a Mediterranean mountain region more than for an Alpine region.
Related collections
Most cited references18
- Record: found
- Abstract: found
- Article: not found
DnaSP, DNA polymorphism analyses by the coalescent and other methods.
- Record: found
- Abstract: found
- Article: not found
Measuring and testing genetic differentiation with ordered versus unordered alleles.
- Record: found
- Abstract: found
- Article: not found