- Record: found
- Abstract: found
- Article: found
Interactions between autophagy and phytohormone signaling pathways in plants
Read this article at
Abstract
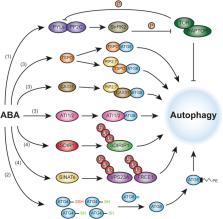
Autophagy is a conserved recycling process with important functions in plant growth, development, and stress responses. Phytohormones also play key roles in the regulation of some of the same processes. Increasing evidence indicates that a close relationship exists between autophagy and phytohormone signaling pathways, and the mechanisms of interaction between these pathways have begun to be revealed. Here, we review recent advances in our understanding of how autophagy regulates hormone signaling and, conversely, how hormones regulate the activity of autophagy, both in plant growth and development and in environmental stress responses. We highlight in particular recent mechanistic insights into the coordination between autophagy and signaling events controlled by the stress hormone abscisic acid and by the growth hormones brassinosteroid and cytokinin and briefly discuss potential connections between autophagy and other phytohormones.
Abstract
This review summarizes emerging evidence for an interplay between plant autophagy and phytohormone pathways, focusing on recent mechanistic insights into autophagy regulation by the stress hormone abscisic acid and by the growth hormones brassinosteroid and cytokinin. Potential connections between autophagy and other phytohormones during plant growth, development, and stress are also briefly discussed.
Related collections
Most cited references149
- Record: found
- Abstract: found
- Article: not found
Abiotic Stress Signaling and Responses in Plants.
- Record: found
- Abstract: found
- Article: not found
Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins.
- Record: found
- Abstract: found
- Article: not found