- Record: found
- Abstract: found
- Article: found
Structural disconnection and functional reorganization in Fabry disease: a multimodal MRI study
Read this article at
Abstract
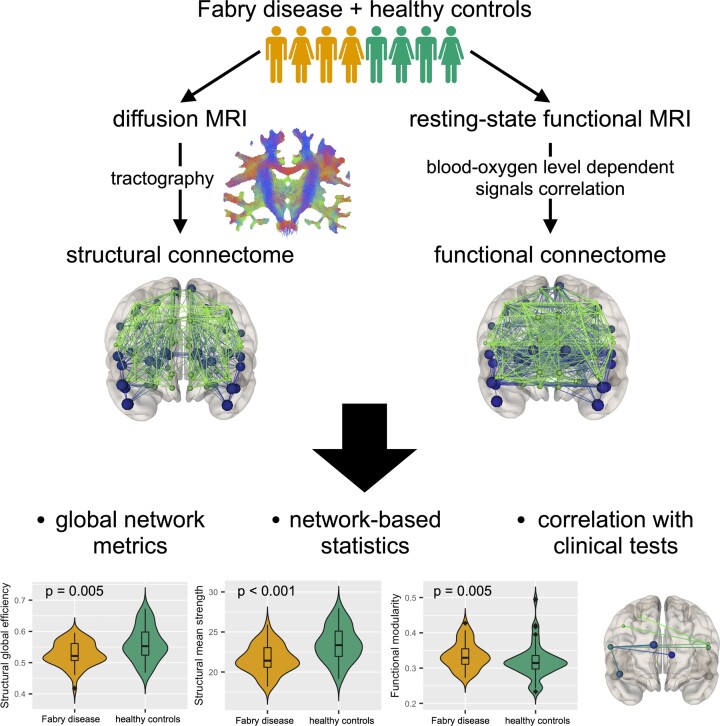
Central nervous system involvement in Fabry disease, a rare systemic X-linked lysosomal storage disorder, is characterized by the presence of heterogeneous but consistent functional and microstructural changes. Nevertheless, knowledge about the degree and extension of macro-scale brain connectivity modifications is to date missing. In this work, we performed connectomic analyses of diffusion and resting-state functional MRI to investigate changes of both structural and functional brain organization in Fabry disease, as well as to explore the relationship between the two and their clinical correlates. In this retrospective cross-sectional study, 46 patients with Fabry disease (28F, 42.2 ± 13.2years) and 49 healthy controls (21F, 42.3 ± 16.3years) were included. All subjects underwent an MRI examination including anatomical, diffusion and resting-state functional sequences. Images were processed to obtain quantitative structural and functional connectomes, where the connections between regions of interest were weighted by the total intra-axonal signal contribution of the corresponding bundle and by the correlation between blood-oxygen level–dependent time series, respectively. We explored between-group differences in terms of both global network properties, expressed with graph measures and specific connected subnetworks, identified using a network-based statistics approach. As exploratory analyses, we also investigated the possible association between cognitive performance and structural and functional connectome modifications at both global and subnetwork level in a subgroup of patients ( n = 11). Compared with healthy controls, patients with Fabry disease showed a significantly reduced global efficiency ( P = 0.005) and mean strength ( P < 0.001) in structural connectomes, together with an increased modularity ( P = 0.005) in functional networks. As for the network-based statistics analysis, a subnetwork with decreased structural connectivity in patients with Fabry disease compared with healthy controls emerged, with eight nodes mainly located at the level of frontal or deep grey-matter areas. When probing the relation between altered global network metrics and neuropsychological tests, correlations emerged between the structural and functional disruption with results at verbal and working memory tests, respectively. Furthermore, structural disruption at subnetwork level was associated with worse executive functioning, with a significant moderation effect of functional changes suggesting a compensation mechanism. Taken together, these results further expand the current knowledge about brain involvement in Fabry disease, showing widespread structural disconnection and functional reorganization, primarily sustained by loss in axonal integrity and correlating with cognitive performance.
Abstract
Gabusi et al. examined structural and functional changes of brain connectivity in Fabry disease using diffusion and resting-state functional MRI data. The connectome analysis of brain networks revealed that patients with Fabry disease show both a structural disconnection (due to mild but widespread axonal damage) and a functional reorganization, associated to cognitive performances.
Graphical Abstract
Related collections
Most cited references70
- Record: found
- Abstract: found
- Article: not found
Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain.
- Record: found
- Abstract: found
- Article: not found
The organization of the human cerebral cortex estimated by intrinsic functional connectivity.
- Record: found
- Abstract: found
- Article: not found