- Record: found
- Abstract: found
- Article: found
Molecular and cytological profiling of biological aging of mouse cochlear inner and outer hair cells
Read this article at
SUMMARY
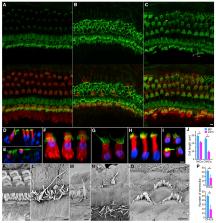
Age-related hearing loss (ARHL) negatively impacts quality of life in the elderly population. The prevalent cause of ARHL is loss of mechanosensitive cochlear hair cells (HCs). The molecular and cellular mechanisms of HC degeneration remain poorly understood. Using RNA-seq transcriptomic analyses of inner and outer HCs isolated from young and aged mice, we show that HC aging is associated with changes in key molecular processes, including transcription, DNA damage, autophagy, and oxidative stress, as well as genes related to HC specialization. At the cellular level, HC aging is characterized by loss of stereocilia, shrinkage of HC soma, and reduction in outer HC mechanical properties, suggesting that functional decline in mechanotransduction and cochlear amplification precedes HC loss and contributes to ARHL. Our study reveals molecular and cytological profiles of aging HCs and identifies genes such as Sod1, Sirt6, Jund, and Cbx3 as biomarkers and potential therapeutic targets for ameliorating ARHL.
Graphical abstract

In brief
Using RNA-seq, advanced imaging, and electrophysiology, Liu et al. reveal molecular and cytological profiles of aging cochlear hair cells. Their study also suggests that a functional decline in mechanotransduction and cochlear amplification precedes hair cell loss and contributes to age-related hearing loss.
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: found
The Hallmarks of Aging

- Record: found
- Abstract: found
- Article: found
ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks
- Record: found
- Abstract: found
- Article: not found