- Record: found
- Abstract: found
- Article: found
ADAR1-mediated RNA editing links ganglioside catabolism to glioblastoma stem cell maintenance

Read this article at
Abstract
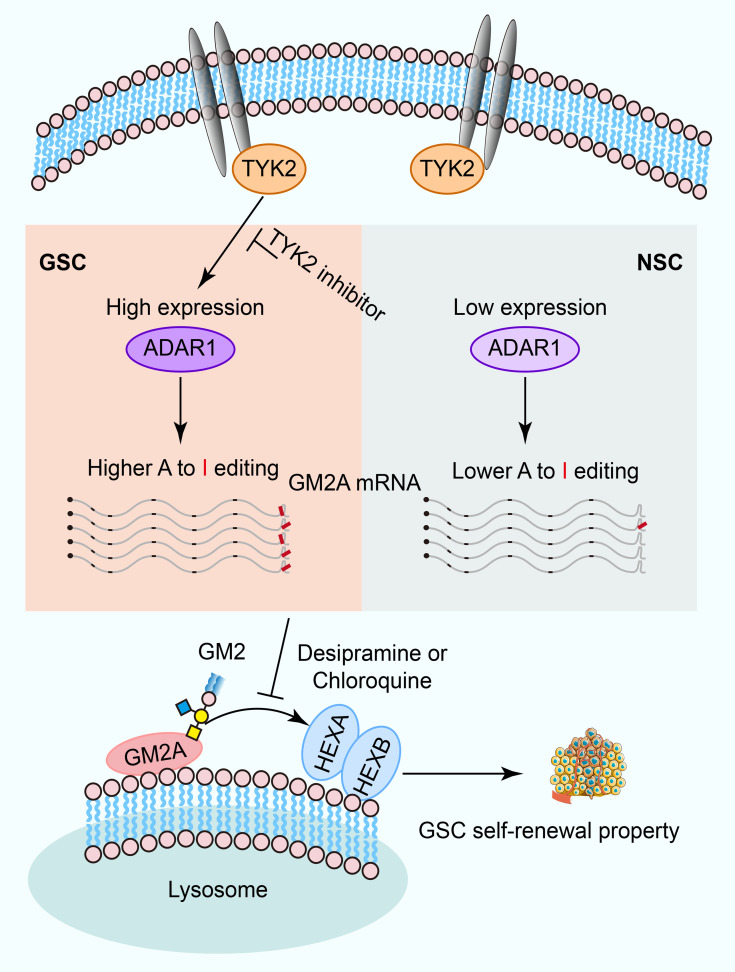
Glioblastoma (GBM) is the most common and lethal primary malignant brain tumor, containing GBM stem cells (GSCs) that contribute to therapeutic resistance and relapse. Exposing potential GSC vulnerabilities may provide therapeutic strategies against GBM. Here, we interrogated the role of adenosine-to-inosine (A-to-I) RNA editing mediated by adenosine deaminase acting on RNA 1 (ADAR1) in GSCs and found that both ADAR1 and global RNA editomes were elevated in GSCs compared with normal neural stem cells. ADAR1 inactivation or blocking of the upstream JAK/STAT pathway through TYK2 inhibition impaired GSC self-renewal and stemness. Downstream of ADAR1, RNA editing of the 3′-UTR of GM2A, a key ganglioside catabolism activator, proved to be critical, as interference with ganglioside catabolism and disruption of ADAR1 showed a similar functional impact on GSCs. These findings reveal that RNA editing links ganglioside catabolism to GSC self-renewal and stemness, exposing a potential vulnerability of GBM for therapeutic intervention.
Abstract
Related collections
Most cited references97

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found