- Record: found
- Abstract: found
- Article: found
IPDfromKM: reconstruct individual patient data from published Kaplan-Meier survival curves

Read this article at
Abstract
Background
When applying secondary analysis on published survival data, it is critical to obtain each patient’s raw data, because the individual patient data (IPD) approach has been considered as the gold standard of data analysis. However, researchers often lack access to IPD. We aim to propose a straightforward and robust approach to obtain IPD from published survival curves with a user-friendly software platform.
Results
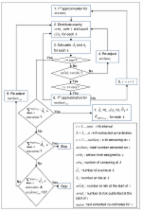
Improving upon existing methods, we propose an easy-to-use, two-stage approach to reconstruct IPD from published Kaplan-Meier (K-M) curves. Stage 1 extracts raw data coordinates and Stage 2 reconstructs IPD using the proposed method. To facilitate the use of the proposed method, we developed the R package IPDfromKM and an accompanying web-based Shiny application. Both the R package and Shiny application have an “all-in-one” feature such that users can use them to extract raw data coordinates from published K-M curves, reconstruct IPD from the extracted data coordinates, visualize the reconstructed IPD, assess the accuracy of the reconstruction, and perform secondary analysis on the basis of the reconstructed IPD. We illustrate the use of the R package and the Shiny application with K-M curves from published studies. Extensive simulations and real-world data applications demonstrate that the proposed method has high accuracy and great reliability in estimating the number of events, number of patients at risk, survival probabilities, median survival times, and hazard ratios.
Conclusions
IPDfromKM has great flexibility and accuracy to reconstruct IPD from published K-M curves with different shapes. We believe that the R package and the Shiny application will greatly facilitate the potential use of quality IPD and advance the use of secondary data to facilitate informed decision making in medical research.
Related collections
Most cited references12
- Record: found
- Abstract: not found
- Article: not found
Nonparametric Estimation from Incomplete Observations
- Record: found
- Abstract: found
- Article: not found
Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial.
- Record: found
- Abstract: found
- Article: found