- Record: found
- Abstract: found
- Article: found
Impaired SorLA maturation and trafficking as a new mechanism for SORL1 missense variants in Alzheimer disease
Read this article at
Abstract
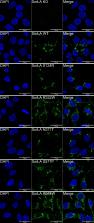
The SorLA protein, encoded by the SORL1 gene, is a major player in Alzheimer’s disease (AD) pathophysiology. Functional and genetic studies demonstrated that SorLA deficiency results in increased production of Aβ peptides, and thus a higher risk of AD. A large number of SORL1 missense variants have been identified in AD patients, but their functional consequences remain largely undefined. Here, we identified a new pathophysiological mechanism, by which rare SORL1 missense variants identified in AD patients result in altered maturation and trafficking of the SorLA protein. An initial screening, based on the overexpression of 70 SorLA variants in HEK293 cells, revealed that 15 of them (S114R, R332W, G543E, S564G, S577P, R654W, R729W, D806N, Y934C, D1535N, D1545E, P1654L, Y1816C, W1862C, P1914S) induced a maturation and trafficking-deficient phenotype. Three of these variants (R332W, S577P, and R654W) and two maturation-competent variants (S124R and N371T) were further studied in details in CRISPR/Cas9-modified hiPSCs. When expressed at endogenous levels, the R332W, S577P, and R654W SorLA variants also showed a maturation defective profile. We further demonstrated that these variants were largely retained in the endoplasmic reticulum, resulting in a reduction in the delivery of SorLA mature protein to the plasma membrane and to the endosomal system. Importantly, expression of the R332W and R654W variants in hiPSCs was associated with a clear increase of Aβ secretion, demonstrating a loss-of-function effect of these SorLA variants regarding this ultimate readout, and a direct link with AD pathophysiology. Furthermore, structural analysis of the impact of missense variants on SorLA protein suggested that impaired cellular trafficking of SorLA protein could be due to subtle variations of the protein 3D structure resulting from changes in the interatomic interactions.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.

- Record: found
- Abstract: found
- Article: found